| schema location: | https://github.com/bio-tools/biotoolsSchema/blob/master/stable/biotools.xsd |
| attributeFormDefault: | unqualified |
| elementFormDefault: | qualified |
| targetNamespace: | biotoolsSchema |
| Elements | Complex types | Simple types |
| dataType | biotoolsIdType | |
| pmcid | EDAMdata | doiType |
| pmid | EDAMformat | enumType |
| tool | linkType | nameType |
| tools | ontologyConcept | textType |
| url | urlftpType | |
| urlType | ||
| versionType |
element email
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | restriction of xs:token | |||||||||
| properties |
|
|||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="email"> <xs:annotation> <xs:documentation>Email address of the entity that is credited.</xs:documentation> <xs:appinfo> <uiTip>Email address of credited entity.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="joebloggst@elixir-dk.org"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:whiteSpace value="collapse"/> <xs:pattern value="[A-Za-z0-9_]+([-+.'][A-Za-z0-9_]+)*@[A-Za-z0-9_]+([-.][A-Za-z0-9_]+)*\.[A-Za-z0-9_]+([-.][A-Za-z0-9_]+)*"/> </xs:restriction> </xs:simpleType> </xs:element> |
element pmcid
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:token | ||||||||
| properties |
|
||||||||
| used by |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="pmcid"> <xs:annotation> <xs:documentation>PubMed Central Identifier (PMCID) of a publication about the software.</xs:documentation> <xs:appinfo>PubMed Central Identifier.</xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="PMC4702812"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>PubMed Central Identifier.</uiTip> > </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="(PMC)[1-9][0-9]{0,8}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element pmid
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of xs:token | ||||||
| properties |
|
||||||
| used by |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="pmid"> <xs:annotation> <xs:appinfo> <uiTip>PubMed Identifier.</uiTip> > </xs:appinfo> <xs:documentation>PubMed Identifier (PMID) of a publication about the software.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="26538599"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="[1-9][0-9]{0,8}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool
| diagram | 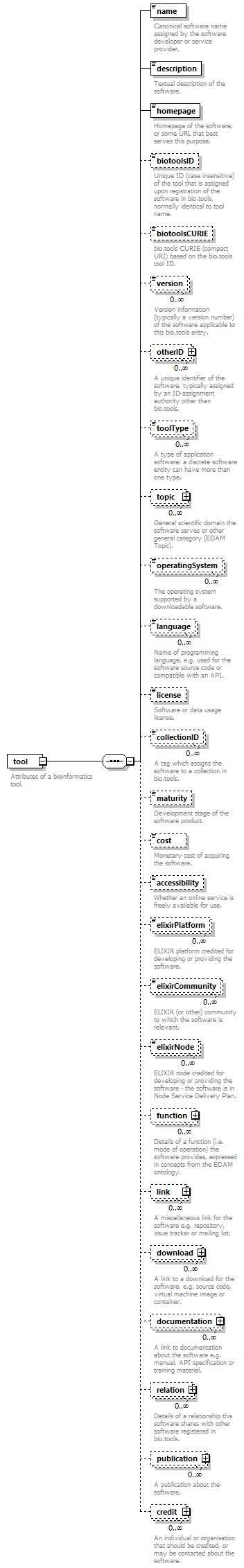 |
||
| namespace | biotoolsSchema | ||
| properties |
|
||
| children | name description homepage biotoolsID biotoolsCURIE version otherID toolType topic operatingSystem language license collectionID maturity cost accessibility elixirPlatform elixirCommunity elixirNode function link download documentation relation publication credit | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:element name="tool"> <xs:annotation> <xs:documentation>Attributes of a bioinformatics tool.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="name" type="nameType"> <xs:annotation> <xs:documentation>Canonical software name assigned by the software developer or service provider.</xs:documentation> <xs:appinfo> <uiTip>Software name.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="needle"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="description"> <xs:annotation> <xs:documentation>Textual description of the software.</xs:documentation> <xs:documentation>This can be a few sentences copy-pasted from the software homepage.</xs:documentation> <xs:appinfo> <uiTip>Software description, e.g. a few sentences adapted from the software publication abstract or homepage.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="needle reads two input sequences and writes their optimal global sequence alignment to file. It uses the Needleman-Wunsch alignment algorithm to find the optimum alignment (including gaps) of two sequences along their entire length. The algorithm uses a dynamic programming method to ensure the alignment is optimum, by exploring all possible alignments and choosing the best."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="textType"> <xs:maxLength value="1000"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="homepage" type="urlftpType"> <xs:annotation> <xs:documentation>Homepage of the software, or some URL that best serves this purpose.</xs:documentation> <xs:appinfo> <uiTip>Homepage of the software, or some URL that best serves this purpose.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="http://emboss.open-bio.org/rel/rel6/apps/needle.html"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="biotoolsID" type="biotoolsIdType" minOccurs="0"> <xs:annotation> <xs:documentation>Unique ID (case insensitive) of the tool that is assigned upon registration of the software in bio.tools, normally identical to tool name.</xs:documentation> <xs:appinfo> <uiTip>Unique ID of the tool that is assigned upon registration of the software in bio.tools.</uiTip> </xs:appinfo> <xs:documentation>bio.tools IDs are set by bio.tools admin and will be disregarded if specified in a payload (e.g. PUSH, POST) to the bio.tools API.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="biotoolsCURIE" minOccurs="0"> <xs:annotation> <xs:documentation>bio.tools CURIE (compact URI) based on the bio.tools tool ID.</xs:documentation> <xs:documentation>The bio.tools CURIE is simply the bio.tools tool ID with the prefix "biotools:".</xs:documentation> <xs:appinfo> <uiTip>bio.tools CURIE (compact URI) based on the bio.tools tool ID.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="biotools:signalp"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>bio.tools CURIEs are set by bio.tools admin and will be disregarded if specified in a payload (e.g. PUSH, POST) to the bio.tools API.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="biotools:[_\-.0-9a-zA-Z]*"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="version" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this bio.tools entry.</xs:documentation> <xs:appinfo> <uiTip>Version information (typically a version number) of the software applicable to this bio.tools entry.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="versionType"/> </xs:simpleType> </xs:element> <xs:element name="otherID" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A unique identifier of the software, typically assigned by an ID-assignment authority other than bio.tools.</xs:documentation> <xs:appinfo> <uiTip>Unique identifier of the software not (typically) assigned by bio.tools.</uiTip> </xs:appinfo> <xs:documentation>This field is not normally used for bio.tools toolIDs, but could be in the exceptional case that multiple such IDs were needed for a given entry.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="value"> <xs:annotation> <xs:documentation>Value of tool identifier.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.1038/nmeth.1701"/> <altova:example value="RRID:SCR_001156"/> <altova:example value="cpe:2.3:a:microsoft:internet_explorer:8.0.6001:beta:*:*:*:*:*:*"/> <altova:example value="biotools:signalp"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>Value of tool identifier.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="10\.[0-9]{4,9}/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> <xs:pattern value="(rrid|RRID):.+"/> <xs:pattern value="(cpe|CPE):.+"/> <xs:pattern value="(BIOTOOLS|biotools):[_\-.0-9a-zA-Z]*"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="type" minOccurs="0"> <xs:annotation> <xs:documentation>Type of tool identifier.</xs:documentation> <xs:appinfo> <uiTip>Type of tool identifier.</uiTip> <enum> <enumItem> <term>doi</term> <uiTip>Digital Object Identifier of the software assigned (typically) by the software developer or service provider.</uiTip> </enumItem> <enumItem> <term>rrid</term> <uiTip>Research Resource Identifier as used by the NIH-supported Resource Identification Portal (https://scicrunch.org/resources).</uiTip> </enumItem> <enumItem> <term>cpe</term> <uiTip>Common Platform Enumeration (CPE) identifier as listed in the CPE dictionary (https://cpe.mitre.org/dictionary/).</uiTip> </enumItem> <enumItem> <term>biotoolsCURIE</term> <uiTip>bio.tools CURIE (secondary identifier).</uiTip> </enumItem> </enum> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="doi"/> <altova:example value="rrid"/> <altova:example value="cpe"/> <altova:example value="biotoolsCURIE"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="doi"/> <xs:enumeration value="rrid"/> <xs:enumeration value="cpe"/> <xs:enumeration value="biotoolsCURIE"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="version" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this identifier.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="versionType"/> </xs:simpleType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="toolType" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A type of application software: a discrete software entity can have more than one type.</xs:documentation> <xs:documentation>bio.tools includes all types of bioinformatics tools: application software with well-defined data processing functions (inputs, outputs and operations). When registering a tool, one or more tool types may be assigned, reflecting the different facets of the software being described.</xs:documentation> <xs:appinfo> <uiTip>Type of application software. A tool may have more than one type.</uiTip> > <enum> <enumItem> <term>Bioinformatics portal</term> <uiTip>A web site providing a platform/portal to multiple resources used for research in a focused area, including biological databases, web applications, training resources and so on.</uiTip> </enumItem> <enumItem> <term>Command-line tool</term> <uiTip>A tool with a text-based (command-line) interface.</uiTip> </enumItem> <enumItem> <term>Database portal</term> <uiTip>A Web site providing a portal to a biological database, typically allowing a user to browse, deposit, search, visualise, analyse or download data.</uiTip> </enumItem> <enumItem> <term>Desktop application</term> <uiTip>A tool with a graphical user interface that runs on your desktop environment, e.g. on a PC or mobile device.</uiTip> </enumItem> <enumItem> <term>Library</term> <uiTip>A collection of components that are used to construct other tools. bio.tools scope includes component libraries performing high-level bioinformatics functions but excludes lower-level programming libraries.</uiTip> </enumItem> <enumItem> <term>Ontology</term> <uiTip>A collection of information about concepts, including terms, synonyms, descriptions etc.</uiTip> </enumItem> <enumItem> <term>Plug-in</term> <uiTip>A software component encapsulating a set of related functions, which are not standalone but depend upon (and typically extend the function of) other software for its use, e.g. a JavaScript widget, plug-in, extension or add-on.</uiTip> </enumItem> <enumItem> <term>Script</term> <uiTip>A tool written for some run-time environment (e.g. other applications or an OS shell) that automates the execution of tasks. Often a small program written in a general-purpose languages (e.g. Perl, Python) or some domain-specific languages (e.g. sed).</uiTip> </enumItem> <enumItem> <term>SPARQL endpoint</term> <uiTip>A service that provides queries over an RDF knowledge base via the SPARQL query language and protocol, and returns results via HTTP.</uiTip> </enumItem> <enumItem> <term>Suite</term> <uiTip>A collection of tools which are bundled together into a convenient toolkit. Such tools typically share related functionality, a common user interface and can exchange data conveniently. This includes collections of stand-alone command-line tools, or Web applications within a common portal.</uiTip> </enumItem> <enumItem> <term>Web application</term> <uiTip>A tool with a graphical user interface that runs in your Web browser.</uiTip> </enumItem> <enumItem> <term>Web API</term> <uiTip>An application programming interface (API) consisting of endpoints to a request-response message system accessible via HTTP. Includes everything from simple data-access URLs to RESTful APIs.</uiTip> </enumItem> <enumItem> <term>Web service</term> <uiTip>An API described in a machine readable form (typically WSDL) providing programmatic access via SOAP over HTTP.</uiTip> </enumItem> <enumItem> <term>Workbench</term> <uiTip>An application or suite with a graphical user interface, providing an integrated environment for data analysis which includes or may be extended with any number of functions or tools. Includes workflow systems, platforms, frameworks etc.</uiTip> </enumItem> <enumItem> <term>Workflow</term> <uiTip>A set of tools which have been composed together into a pipeline of some sort. Such tools are (typically) standalone, but are composed for convenience, for instance for batch execution via some workflow engine or script.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Bioinformatics portal"/> <xs:enumeration value="Command-line tool"/> <xs:enumeration value="Database portal"/> <xs:enumeration value="Desktop application"/> <xs:enumeration value="Library"/> <xs:enumeration value="Ontology"/> <xs:enumeration value="Plug-in"/> <xs:enumeration value="Script"/> <xs:enumeration value="SPARQL endpoint"/> <xs:enumeration value="Suite"/> <xs:enumeration value="Web application"/> <xs:enumeration value="Web API"/> <xs:enumeration value="Web service"/> <xs:enumeration value="Workbench"/> <xs:enumeration value="Workflow"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="topic" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>General scientific domain the software serves or other general category (EDAM Topic).</xs:documentation> <xs:documentation>An EDAM Topic concept URL and / or term are specified, e.g. "Proteomics", http://edamontology.org/topic_0121.</xs:documentation> <xs:appinfo> <uiTip>Scientific topic (EDAM Topic).</uiTip> > </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Topic concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Topic URL.</uiTip> > </xs:appinfo> <xs:documentation>The URL must be in the EDAM Topic namespace, i.e. http://edamontology.org/topic_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/topic_0121"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/topic_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Topic term (preferred label or synonym).</xs:documentation> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <uiTip>EDAM Topic term (preferred label or synonym).</uiTip> > </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Proteomics"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="operatingSystem" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The operating system supported by a downloadable software.</xs:documentation> <xs:appinfo> <uiTip>Operating system supported by a downloadable software package.</uiTip> <enum> <enumItem> <term>Linux</term> <uiTip>All flavours of Linux/UNIX operating systems.</uiTip> </enumItem> <enumItem> <term>Windows</term> <uiTip>All flavours of Microsoft Windows operating system.</uiTip> </enumItem> <enumItem> <term>Mac</term> <uiTip>All flavours of Apple Macintosh operating systems (primarily Mac OS X).</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Linux"/> <xs:enumeration value="Windows"/> <xs:enumeration value="Mac"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="language" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Name of programming language, e.g. used for the software source code or compatible with an API.</xs:documentation> <xs:appinfo> <uiTip>Source code language.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="ActionScript"/> <xs:enumeration value="Ada"/> <xs:enumeration value="AppleScript"/> <xs:enumeration value="Assembly language"/> <xs:enumeration value="AWK"/> <xs:enumeration value="Bash"/> <xs:enumeration value="C"/> <xs:enumeration value="C#"/> <xs:enumeration value="C++"/> <xs:enumeration value="COBOL"/> <xs:enumeration value="ColdFusion"/> <xs:enumeration value="CWL"/> <xs:enumeration value="D"/> <xs:enumeration value="Delphi"/> <xs:enumeration value="Dylan"/> <xs:enumeration value="Eiffel"/> <xs:enumeration value="Elm"/> <xs:enumeration value="Forth"/> <xs:enumeration value="Fortran"/> <xs:enumeration value="Groovy"/> <xs:enumeration value="Haskell"/> <xs:enumeration value="Icarus"/> <xs:enumeration value="Java"/> <xs:enumeration value="JavaScript"/> <xs:enumeration value="Julia"/> <xs:enumeration value="JSP"/> <xs:enumeration value="LabVIEW"/> <xs:enumeration value="Lisp"/> <xs:enumeration value="Lua"/> <xs:enumeration value="Maple"/> <xs:enumeration value="Mathematica"/> <xs:enumeration value="MATLAB"/> <xs:enumeration value="MLXTRAN"/> <xs:enumeration value="NMTRAN"/> <xs:enumeration value="OCaml"/> <xs:enumeration value="Pascal"/> <xs:enumeration value="Perl"/> <xs:enumeration value="PHP"/> <xs:enumeration value="Prolog"/> <xs:enumeration value="PyMOL"/> <xs:enumeration value="Python"/> <xs:enumeration value="R"/> <xs:enumeration value="Racket"/> <xs:enumeration value="REXX"/> <xs:enumeration value="Ruby"/> <xs:enumeration value="SAS"/> <xs:enumeration value="Scala"/> <xs:enumeration value="Scheme"/> <xs:enumeration value="Shell"/> <xs:enumeration value="Smalltalk"/> <xs:enumeration value="SQL"/> <xs:enumeration value="Turing"/> <xs:enumeration value="Verilog"/> <xs:enumeration value="VHDL"/> <xs:enumeration value="Visual Basic"/> <xs:enumeration value="XAML"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="license" minOccurs="0"> <xs:annotation> <xs:documentation>Software or data usage license.</xs:documentation> <xs:appinfo> <uiTip>Software or data usage license. For online services, this refers to the license of the underlying software or data.</uiTip> </xs:appinfo> <xs:documentation>Identifier from the SPDX license list (https://spdx.org/licenses/). In future, based on the supplied license, a label e.g. "Open-source" may be applied to the entry in bio.tools.</xs:documentation> <xs:appinfo> <enum> <enumItem> <term>Open-source</term> <uiTip>Software is made available under a license approved by the Open Source Initiative (OSI). The software is distributed in a way that satisfies the 10 criteria of the Open Source Definition maintained by OSI (see https://opensource.org/docs/osd). The source code is available to others.</uiTip> </enumItem> <enumItem> <term>Free software</term> <uiTip>Free as in “freedom” not necessarily free of charge. Software is made available under a license approved by the Free Software Foundation (FSF). The software satisfies the criteria of the Free Software Definition maintained by FSF (see http://www.gnu.org/philosophy/free-sw.html). The source code is available to others.</uiTip> </enumItem> <enumItem> <term>Free and open source</term> <uiTip>Software is made available under a license approved by both the Open Source Initiative (OSI) and the Free Software Foundation (FSF), and satisfies the criteria of the OSI Open Source Definition maintained (https://opensource.org/docs/osd) and the FSF Free Software Definition (http://www.gnu.org/philosophy/free-sw.html). Such software ensures users have the freedom to run, copy, distribute, study, change and improve the software. The source code is available to others.</uiTip> </enumItem> <enumItem> <term>Copyleft</term> <uiTip>Software is made available under a license designated as “copyleft” by the Free Software Foundation (FSF). The license ensures such software is free and that all modified and extended versions of the program are free as well. Free as in “freedom” not necessarily free of charge, as per the Free Software Definition maintained by FSF (see http://www.gnu.org/philosophy/free-sw.html).</uiTip> </enumItem> <enumItem> <term>Proprietary</term> <uiTip>Software for which the software's publisher or another person retains intellectual property rights—usually copyright of the source code, but sometimes patent rights.</uiTip> </enumItem> </enum> <enumItem> <term>Freeware</term> <uiTip>Proprietary software that is available for use at no monetary cost. In other words, freeware may be used without payment but may usually not be modified, re-distributed or reverse-engineered without the author's permission.</uiTip> </enumItem> <enumItem> <term>Not licensed</term> <uiTip>Software which is not licensed and is not proprietary.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Software under license not currently supported by biotoolsSchema.</uiTip> </enumItem> </xs:appinfo> <xs:documentation>Use "Proprietary" where the software must be obtained from the provider (e.g. for money), and is then owned. Use "Freeware" for proprietary software that is available at no monetary cost. Use "Not licensed" for software which is not licensed and is not proprietary, and "Other" for software under license not currently supported by biotoolsSchema.</xs:documentation> <xs:documentation>Note that for online services, the license attribute refers to the license of the underlying software or data, or a part of those.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="0BSD"/> <xs:enumeration value="AAL"/> <xs:enumeration value="ADSL"/> <xs:enumeration value="AFL-1.1"/> <xs:enumeration value="AFL-1.2"/> <xs:enumeration value="AFL-2.0"/> <xs:enumeration value="AFL-2.1"/> <xs:enumeration value="AFL-3.0"/> <xs:enumeration value="AGPL-1.0"/> <xs:enumeration value="AGPL-3.0"/> <xs:enumeration value="AMDPLPA"/> <xs:enumeration value="AML"/> <xs:enumeration value="AMPAS"/> <xs:enumeration value="ANTLR-PD"/> <xs:enumeration value="APAFML"/> <xs:enumeration value="APL-1.0"/> <xs:enumeration value="APSL-1.0"/> <xs:enumeration value="APSL-1.1"/> <xs:enumeration value="APSL-1.2"/> <xs:enumeration value="APSL-2.0"/> <xs:enumeration value="Abstyles"/> <xs:enumeration value="Adobe-2006"/> <xs:enumeration value="Adobe-Glyph"/> <xs:enumeration value="Afmparse"/> <xs:enumeration value="Aladdin"/> <xs:enumeration value="Apache-1.0"/> <xs:enumeration value="Apache-1.1"/> <xs:enumeration value="Apache-2.0"/> <xs:enumeration value="Artistic-1.0"/> <xs:enumeration value="Artistic-1.0-Perl"/> <xs:enumeration value="Artistic-1.0-cl8"/> <xs:enumeration value="Artistic-2.0"/> <xs:enumeration value="BSD-2-Clause"/> <xs:enumeration value="BSD-2-Clause-FreeBSD"/> <xs:enumeration value="BSD-2-Clause-NetBSD"/> <xs:enumeration value="BSD-3-Clause"/> <xs:enumeration value="BSD-3-Clause-Attribution"/> <xs:enumeration value="BSD-3-Clause-Clear"/> <xs:enumeration value="BSD-3-Clause-LBNL"/> <xs:enumeration value="BSD-3-Clause-No-Nuclear-License"/> <xs:enumeration value="BSD-3-Clause-No-Nuclear-License-2014"/> <xs:enumeration value="BSD-3-Clause-No-Nuclear-Warranty"/> <xs:enumeration value="BSD-4-Clause"/> <xs:enumeration value="BSD-4-Clause-UC"/> <xs:enumeration value="BSD-Protection"/> <xs:enumeration value="BSD-Source-Code"/> <xs:enumeration value="BSL-1.0"/> <xs:enumeration value="Bahyph"/> <xs:enumeration value="Barr"/> <xs:enumeration value="Beerware"/> <xs:enumeration value="BitTorrent-1.0"/> <xs:enumeration value="BitTorrent-1.1"/> <xs:enumeration value="Borceux"/> <xs:enumeration value="CATOSL-1.1"/> <xs:enumeration value="CC-BY-1.0"/> <xs:enumeration value="CC-BY-2.0"/> <xs:enumeration value="CC-BY-2.5"/> <xs:enumeration value="CC-BY-3.0"/> <xs:enumeration value="CC-BY-4.0"/> <xs:enumeration value="CC-BY-NC-1.0"/> <xs:enumeration value="CC-BY-NC-2.0"/> <xs:enumeration value="CC-BY-NC-2.5"/> <xs:enumeration value="CC-BY-NC-3.0"/> <xs:enumeration value="CC-BY-NC-4.0"/> <xs:enumeration value="CC-BY-NC-ND-1.0"/> <xs:enumeration value="CC-BY-NC-ND-2.0"/> <xs:enumeration value="CC-BY-NC-ND-2.5"/> <xs:enumeration value="CC-BY-NC-ND-3.0"/> <xs:enumeration value="CC-BY-NC-ND-4.0"/> <xs:enumeration value="CC-BY-NC-SA-1.0"/> <xs:enumeration value="CC-BY-NC-SA-2.0"/> <xs:enumeration value="CC-BY-NC-SA-2.5"/> <xs:enumeration value="CC-BY-NC-SA-3.0"/> <xs:enumeration value="CC-BY-NC-SA-4.0"/> <xs:enumeration value="CC-BY-ND-1.0"/> <xs:enumeration value="CC-BY-ND-2.0"/> <xs:enumeration value="CC-BY-ND-2.5"/> <xs:enumeration value="CC-BY-ND-3.0"/> <xs:enumeration value="CC-BY-ND-4.0"/> <xs:enumeration value="CC-BY-SA-1.0"/> <xs:enumeration value="CC-BY-SA-2.0"/> <xs:enumeration value="CC-BY-SA-2.5"/> <xs:enumeration value="CC-BY-SA-3.0"/> <xs:enumeration value="CC-BY-SA-4.0"/> <xs:enumeration value="CC0-1.0"/> <xs:enumeration value="CDDL-1.0"/> <xs:enumeration value="CDDL-1.1"/> <xs:enumeration value="CECILL-1.0"/> <xs:enumeration value="CECILL-1.1"/> <xs:enumeration value="CECILL-2.0"/> <xs:enumeration value="CECILL-2.1"/> <xs:enumeration value="CECILL-B"/> <xs:enumeration value="CECILL-C"/> <xs:enumeration value="CNRI-Jython"/> <xs:enumeration value="CNRI-Python"/> <xs:enumeration value="CNRI-Python-GPL-Compatible"/> <xs:enumeration value="CPAL-1.0"/> <xs:enumeration value="CPL-1.0"/> <xs:enumeration value="CPOL-1.02"/> <xs:enumeration value="CUA-OPL-1.0"/> <xs:enumeration value="Caldera"/> <xs:enumeration value="ClArtistic"/> <xs:enumeration value="Condor-1.1"/> <xs:enumeration value="Crossword"/> <xs:enumeration value="CrystalStacker"/> <xs:enumeration value="Cube"/> <xs:enumeration value="D-FSL-1.0"/> <xs:enumeration value="DOC"/> <xs:enumeration value="DSDP"/> <xs:enumeration value="Dotseqn"/> <xs:enumeration value="ECL-1.0"/> <xs:enumeration value="ECL-2.0"/> <xs:enumeration value="EFL-1.0"/> <xs:enumeration value="EFL-2.0"/> <xs:enumeration value="EPL-1.0"/> <xs:enumeration value="EUDatagrid"/> <xs:enumeration value="EUPL-1.0"/> <xs:enumeration value="EUPL-1.1"/> <xs:enumeration value="Entessa"/> <xs:enumeration value="ErlPL-1.1"/> <xs:enumeration value="Eurosym"/> <xs:enumeration value="FSFAP"/> <xs:enumeration value="FSFUL"/> <xs:enumeration value="FSFULLR"/> <xs:enumeration value="FTL"/> <xs:enumeration value="Fair"/> <xs:enumeration value="Frameworx-1.0"/> <xs:enumeration value="FreeImage"/> <xs:enumeration value="GFDL-1.1"/> <xs:enumeration value="GFDL-1.2"/> <xs:enumeration value="GFDL-1.3"/> <xs:enumeration value="GL2PS"/> <xs:enumeration value="GPL-1.0"/> <xs:enumeration value="GPL-2.0"/> <xs:enumeration value="GPL-3.0"/> <xs:enumeration value="Giftware"/> <xs:enumeration value="Glide"/> <xs:enumeration value="Glulxe"/> <xs:enumeration value="HPND"/> <xs:enumeration value="HaskellReport"/> <xs:enumeration value="IBM-pibs"/> <xs:enumeration value="ICU"/> <xs:enumeration value="IJG"/> <xs:enumeration value="IPA"/> <xs:enumeration value="IPL-1.0"/> <xs:enumeration value="ISC"/> <xs:enumeration value="ImageMagick"/> <xs:enumeration value="Imlib2"/> <xs:enumeration value="Info-ZIP"/> <xs:enumeration value="Intel"/> <xs:enumeration value="Intel-ACPI"/> <xs:enumeration value="Interbase-1.0"/> <xs:enumeration value="JSON"/> <xs:enumeration value="JasPer-2.0"/> <xs:enumeration value="LAL-1.2"/> <xs:enumeration value="LAL-1.3"/> <xs:enumeration value="LGPL-2.0"/> <xs:enumeration value="LGPL-2.1"/> <xs:enumeration value="LGPL-3.0"/> <xs:enumeration value="LGPLLR"/> <xs:enumeration value="LPL-1.0"/> <xs:enumeration value="LPL-1.02"/> <xs:enumeration value="LPPL-1.0"/> <xs:enumeration value="LPPL-1.1"/> <xs:enumeration value="LPPL-1.2"/> <xs:enumeration value="LPPL-1.3a"/> <xs:enumeration value="LPPL-1.3c"/> <xs:enumeration value="Latex2e"/> <xs:enumeration value="Leptonica"/> <xs:enumeration value="LiLiQ-P-1.1"/> <xs:enumeration value="LiLiQ-R-1.1"/> <xs:enumeration value="LiLiQ-Rplus-1.1"/> <xs:enumeration value="Libpng"/> <xs:enumeration value="MIT"/> <xs:enumeration value="MIT-CMU"/> <xs:enumeration value="MIT-advertising"/> <xs:enumeration value="MIT-enna"/> <xs:enumeration value="MIT-feh"/> <xs:enumeration value="MITNFA"/> <xs:enumeration value="MPL-1.0"/> <xs:enumeration value="MPL-1.1"/> <xs:enumeration value="MPL-2.0"/> <xs:enumeration value="MPL-2.0-no-copyleft-exception"/> <xs:enumeration value="MS-PL"/> <xs:enumeration value="MS-RL"/> <xs:enumeration value="MTLL"/> <xs:enumeration value="MakeIndex"/> <xs:enumeration value="MirOS"/> <xs:enumeration value="Motosoto"/> <xs:enumeration value="Multics"/> <xs:enumeration value="Mup"/> <xs:enumeration value="NASA-1.3"/> <xs:enumeration value="NBPL-1.0"/> <xs:enumeration value="NCSA"/> <xs:enumeration value="NGPL"/> <xs:enumeration value="NLOD-1.0"/> <xs:enumeration value="NLPL"/> <xs:enumeration value="NOSL"/> <xs:enumeration value="NPL-1.0"/> <xs:enumeration value="NPL-1.1"/> <xs:enumeration value="NPOSL-3.0"/> <xs:enumeration value="NRL"/> <xs:enumeration value="NTP"/> <xs:enumeration value="Naumen"/> <xs:enumeration value="NetCDF"/> <xs:enumeration value="Newsletr"/> <xs:enumeration value="Nokia"/> <xs:enumeration value="Noweb"/> <xs:enumeration value="Nunit"/> <xs:enumeration value="OCCT-PL"/> <xs:enumeration value="OCLC-2.0"/> <xs:enumeration value="ODbL-1.0"/> <xs:enumeration value="OFL-1.0"/> <xs:enumeration value="OFL-1.1"/> <xs:enumeration value="OGTSL"/> <xs:enumeration value="OLDAP-1.1"/> <xs:enumeration value="OLDAP-1.2"/> <xs:enumeration value="OLDAP-1.3"/> <xs:enumeration value="OLDAP-1.4"/> <xs:enumeration value="OLDAP-2.0"/> <xs:enumeration value="OLDAP-2.0.1"/> <xs:enumeration value="OLDAP-2.1"/> <xs:enumeration value="OLDAP-2.2"/> <xs:enumeration value="OLDAP-2.2.1"/> <xs:enumeration value="OLDAP-2.2.2"/> <xs:enumeration value="OLDAP-2.3"/> <xs:enumeration value="OLDAP-2.4"/> <xs:enumeration value="OLDAP-2.5"/> <xs:enumeration value="OLDAP-2.6"/> <xs:enumeration value="OLDAP-2.7"/> <xs:enumeration value="OLDAP-2.8"/> <xs:enumeration value="OML"/> <xs:enumeration value="OPL-1.0"/> <xs:enumeration value="OSET-PL-2.1"/> <xs:enumeration value="OSL-1.0"/> <xs:enumeration value="OSL-1.1"/> <xs:enumeration value="OSL-2.0"/> <xs:enumeration value="OSL-2.1"/> <xs:enumeration value="OSL-3.0"/> <xs:enumeration value="OpenSSL"/> <xs:enumeration value="PDDL-1.0"/> <xs:enumeration value="PHP-3.0"/> <xs:enumeration value="PHP-3.01"/> <xs:enumeration value="Plexus"/> <xs:enumeration value="PostgreSQL"/> <xs:enumeration value="Python-2.0"/> <xs:enumeration value="QPL-1.0"/> <xs:enumeration value="Qhull"/> <xs:enumeration value="RHeCos-1.1"/> <xs:enumeration value="RPL-1.1"/> <xs:enumeration value="RPL-1.5"/> <xs:enumeration value="RPSL-1.0"/> <xs:enumeration value="RSA-MD"/> <xs:enumeration value="RSCPL"/> <xs:enumeration value="Rdisc"/> <xs:enumeration value="Ruby"/> <xs:enumeration value="SAX-PD"/> <xs:enumeration value="SCEA"/> <xs:enumeration value="SGI-B-1.0"/> <xs:enumeration value="SGI-B-1.1"/> <xs:enumeration value="SGI-B-2.0"/> <xs:enumeration value="SISSL"/> <xs:enumeration value="SISSL-1.2"/> <xs:enumeration value="SMLNJ"/> <xs:enumeration value="SMPPL"/> <xs:enumeration value="SNIA"/> <xs:enumeration value="SPL-1.0"/> <xs:enumeration value="SWL"/> <xs:enumeration value="Saxpath"/> <xs:enumeration value="Sendmail"/> <xs:enumeration value="SimPL-2.0"/> <xs:enumeration value="Sleepycat"/> <xs:enumeration value="Spencer-86"/> <xs:enumeration value="Spencer-94"/> <xs:enumeration value="Spencer-99"/> <xs:enumeration value="SugarCRM-1.1.3"/> <xs:enumeration value="TCL"/> <xs:enumeration value="TMate"/> <xs:enumeration value="TORQUE-1.1"/> <xs:enumeration value="TOSL"/> <xs:enumeration value="UPL-1.0"/> <xs:enumeration value="Unicode-TOU"/> <xs:enumeration value="Unlicense"/> <xs:enumeration value="VOSTROM"/> <xs:enumeration value="VSL-1.0"/> <xs:enumeration value="Vim"/> <xs:enumeration value="W3C"/> <xs:enumeration value="W3C-19980720"/> <xs:enumeration value="WTFPL"/> <xs:enumeration value="Watcom-1.0"/> <xs:enumeration value="Wsuipa"/> <xs:enumeration value="X11"/> <xs:enumeration value="XFree86-1.1"/> <xs:enumeration value="XSkat"/> <xs:enumeration value="Xerox"/> <xs:enumeration value="Xnet"/> <xs:enumeration value="YPL-1.0"/> <xs:enumeration value="YPL-1.1"/> <xs:enumeration value="ZPL-1.1"/> <xs:enumeration value="ZPL-2.0"/> <xs:enumeration value="ZPL-2.1"/> <xs:enumeration value="Zed"/> <xs:enumeration value="Zend-2.0"/> <xs:enumeration value="Zimbra-1.3"/> <xs:enumeration value="Zimbra-1.4"/> <xs:enumeration value="Zlib"/> <xs:enumeration value="bzip2-1.0.5"/> <xs:enumeration value="bzip2-1.0.6"/> <xs:enumeration value="curl"/> <xs:enumeration value="diffmark"/> <xs:enumeration value="dvipdfm"/> <xs:enumeration value="eGenix"/> <xs:enumeration value="gSOAP-1.3b"/> <xs:enumeration value="gnuplot"/> <xs:enumeration value="iMatix"/> <xs:enumeration value="libtiff"/> <xs:enumeration value="mpich2"/> <xs:enumeration value="psfrag"/> <xs:enumeration value="psutils"/> <xs:enumeration value="xinetd"/> <xs:enumeration value="xpp"/> <xs:enumeration value="zlib-acknowledgement"/> <xs:enumeration value="Proprietary"/> <xs:enumeration value="Other"/> <xs:enumeration value="Not licensed"/> <xs:enumeration value="Freeware"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="collectionID" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A tag which assigns the software to a collection in bio.tools.</xs:documentation> <xs:appinfo> <uiTip>Tag for a collection that the software has been assigned to within bio.tools.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="emboss"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="nameType"/> </xs:simpleType> </xs:element> <xs:element name="maturity" minOccurs="0"> <xs:annotation> <xs:documentation>Development stage of the software product.</xs:documentation> <xs:appinfo> <uiTip>How mature the software product is.</uiTip> <enum> <enumItem> <term>Emerging</term> <uiTip>Nascent or early release software that may not yet be fully featured or stable.</uiTip> </enumItem> <enumItem> <term>Mature</term> <uiTip>Software that is generally considered to fulfill several of the following: secure, reliable, actively maintained, fully featured, proven in production environments, has an active community, and is described or cited in the scientific literature.</uiTip> </enumItem> <enumItem> <term>Legacy</term> <uiTip>Software which is no longer in common use, deprecated by the provider, superseded by other software, replaced by a newer version, is obsolete etc.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Emerging"/> <xs:enumeration value="Mature"/> <xs:enumeration value="Legacy"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="cost" minOccurs="0"> <xs:annotation> <xs:documentation>Monetary cost of acquiring the software.</xs:documentation> <xs:appinfo> <uiTip>Monetary cost of acquiring the software.</uiTip> <enum> <enumItem> <term>Free of charge</term> <uiTip>Software which available for use by all, with full functionality, at no monetary cost to the user.</uiTip> </enumItem> <enumItem> <term>Free of charge (with restrictions)</term> <uiTip>Software which is available for use at no monetary cost to the user, but possibly with limited functionality, usage restrictions, or other limitations.</uiTip> </enumItem> <enumItem> <term>Commercial</term> <uiTip>Software which you have to pay to access.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Free of charge"/> <xs:enumeration value="Free of charge (with restrictions)"/> <xs:enumeration value="Commercial"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="accessibility" minOccurs="0"> <xs:annotation> <xs:documentation>Whether an online service is freely available for use.</xs:documentation> <xs:appinfo> <uiTip>Whether the software is freely available for use.</uiTip> <enum> <enumItem> <term>Open access</term> <uiTip>An online service which is available for use to all, but possibly requiring user accounts / authentication.</uiTip> </enumItem> <enumItem> <term>Restricted access</term> <uiTip>An online service which is available for use to a restricted audience, e.g. members of a specific institute.</uiTip> </enumItem> <enumItem> <term>Proprietary</term> <uiTip>Software for which the software's publisher or another person retains intellectual property rights—usually copyright of the source code, but sometimes patent rights.</uiTip> </enumItem> <enumItem> <term>Freeware</term> <uiTip>Proprietary software that is available for use at no monetary cost. In other words, freeware may be used without payment but may usually not be modified, re-distributed or reverse-engineered without the author's permission.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Open access"/> <xs:enumeration value="Restricted access"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="elixirPlatform" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>ELIXIR platform credited for developing or providing the software.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Training"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>ELIXIR platform credited for developing or providing the software.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Data"/> <xs:enumeration value="Tools"/> <xs:enumeration value="Compute"/> <xs:enumeration value="Interoperability"/> <xs:enumeration value="Training"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="elixirCommunity" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>ELIXIR (or other) community to which the software is relevant.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="3D-BioInfo"/> <xs:enumeration value="Federated Human Data"/> <xs:enumeration value="Galaxy"/> <xs:enumeration value="Human Copy Number Variation"/> <xs:enumeration value="Intrinsically Disordered Proteins"/> <xs:enumeration value="Marine Metagenomics"/> <xs:enumeration value="Metabolomics"/> <xs:enumeration value="Microbial Biotechnology"/> <xs:enumeration value="Plant Sciences"/> <xs:enumeration value="Proteomics"/> <xs:enumeration value="Rare Diseases"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="elixirNode" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>ELIXIR node credited for developing or providing the software - the software is in Node Service Delivery Plan.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="UK"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>ELIXIR node credited for developing or providing the software - the software is in Node Service Delivery Plan.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Belgium"/> <xs:enumeration value="Czech Republic"/> <xs:enumeration value="Denmark"/> <xs:enumeration value="EMBL"/> <xs:enumeration value="Estonia"/> <xs:enumeration value="Finland"/> <xs:enumeration value="France"/> <xs:enumeration value="Germany"/> <xs:enumeration value="Greece"/> <xs:enumeration value="Hungary"/> <xs:enumeration value="Ireland"/> <xs:enumeration value="Israel"/> <xs:enumeration value="Italy"/> <xs:enumeration value="Luxembourg"/> <xs:enumeration value="Netherlands"/> <xs:enumeration value="Norway"/> <xs:enumeration value="Portugal"/> <xs:enumeration value="Slovenia"/> <xs:enumeration value="Spain"/> <xs:enumeration value="Sweden"/> <xs:enumeration value="Switzerland"/> <xs:enumeration value="UK"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="function" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of a function (i.e. mode of operation) the software provides, expressed in concepts from the EDAM ontology.</xs:documentation> <xs:appinfo> <uiTip>Details of a function the software provides, expressed in terms from the EDAM ontology.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="operation" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The basic operation(s) performed by this software function (EDAM Operation).</xs:documentation> <xs:documentation>An EDAM Operation concept URL and / or term are specified, e.g. "Multiple sequence alignment", http://edamontology.org/operation_0492.</xs:documentation> <xs:appinfo> <uiTip>Operation performed (EDAM operation).</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Operation concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation URL.</uiTip> </xs:appinfo> <xs:documentation>The URL must be in the EDAM Operation namespace, i.e. http://edamontology.org/operation_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/operation_0492"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/operation_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Operation term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation term (preferred label or synonym).</uiTip> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Multiple sequence alignment"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="input" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of primary input data.</xs:documentation> <xs:appinfo> <uiTip>Details of primary input data.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="dataType"> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary input data, if any (EDAM data). </xs:documentation> <xs:appinfo> <uiTip>Input data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and / or term are specified, e.g. "Protein sequences", http://edamontology.org/data_2976. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the input data (EDAM Format). </xs:documentation> <xs:appinfo> <uiTip>Input data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and / or term are specified, e.g. "FASTA", http://edamontology.org/format_1929.</xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="output" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of primary output data.</xs:documentation> <xs:appinfo> <uiTip>Details of primary output data.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="dataType"> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary output data, if any (EDAM Data).</xs:documentation> <xs:appinfo> <uiTip>Output data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and (optionally) term are specified, e.g. "Sequence alignment", http://edamontology.org/data_0863.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the output data (EDAM Format).</xs:documentation> <xs:appinfo> <uiTip>Output data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and (optionally) term are specified, e.g. “FASTA-aln”, http://edamontology.org/format_1984.</xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="note" minOccurs="0"> <xs:annotation> <xs:documentation>Concise comment about this function, if not apparent from the software description and EDAM annotations.</xs:documentation> <xs:appinfo> <uiTip>General comment about function.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Concise comment about this function, if not apparent from the software description and EDAM annotations."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="textType"/> </xs:simpleType> </xs:element> <xs:element name="cmd" minOccurs="0"> <xs:annotation> <xs:documentation>Relevant command, command-line fragment or option for executing this function / running the tool in this mode.</xs:documentation> <xs:appinfo> <uiTip>Relevant command, command-line fragment or option for executing this function / running the tool in this mode.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="-s best"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:maxLength value="1000"/> <xs:whiteSpace value="collapse"/> <xs:minLength value="1"/> </xs:restriction> </xs:simpleType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="link" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A miscellaneous link for the software e.g. repository, issue tracker or mailing list.</xs:documentation> <xs:appinfo> <uiTip>A miscellaneous link for the software e.g. repository, issue tracker or mailing list.</uiTip> > </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="linkType"> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>A link of some relevance to the software (URL).</xs:documentation> <xs:documentation> <uiTip>Miscellaneous link.</uiTip> </xs:documentation> </xs:annotation> </xs:element> <xs:element name="type" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The type of data, information or system that is obtained when the link is resolved.</xs:documentation> <xs:appinfo> <uiTip>Type of thing linked to.</uiTip> <enum> <enumItem> <term>Discussion forum</term> <uiTip>Online forum for user discussions about the software.</uiTip> </enumItem> <enumItem> <term>Galaxy service</term> <uiTip>An online service providing the tool through the Galaxy platform.</uiTip> </enumItem> <enumItem> <term>Helpdesk</term> <uiTip>A phone line, web site or email-based system providing help to the end-user of the software.</uiTip> </enumItem> <enumItem> <term>Issue tracker</term> <uiTip>Tracker for software issues, bug reports, feature requests etc.</uiTip> </enumItem> <enumItem> <term>Mailing list</term> <uiTip>Mailing list for the software announcements, discussions, support etc.</uiTip> </enumItem> <enumItem> <term>Mirror</term> <uiTip>Mirror of an (identical) online service.</uiTip> </enumItem> <enumItem> <term>Software catalogue</term> <uiTip>Some registry, catalogue etc. other than bio.tools where the tool is also described..</uiTip> </enumItem> <enumItem> <term>Repository</term> <uiTip>A place where source code, data and other files can be retrieved from, typically via platforms like GitHub which provide version control and other features, or something simpler, e.g. an FTP site.</uiTip> </enumItem> <enumItem> <term>Social media</term> <uiTip>A website used by the software community including social networking sites, discussion and support fora, WIKIs etc.</uiTip> </enumItem> <enumItem> <term>Service</term> <uiTip>An online service (other than Galaxy) that provides access (an interface) to the software.</uiTip> </enumItem> <enumItem> <term>Technical monitoring</term> <uiTip>Information about the technical status of a tool.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Other type of link for software - the default if a more specific type is not available.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Discussion forum"/> <xs:enumeration value="Galaxy service"/> <xs:enumeration value="Helpdesk"/> <xs:enumeration value="Issue tracker"/> <xs:enumeration value="Mailing list"/> <xs:enumeration value="Mirror"/> <xs:enumeration value="Software catalogue"/> <xs:enumeration value="Repository"/> <xs:enumeration value="Service"/> <xs:enumeration value="Social media"/> <xs:enumeration value="Technical monitoring"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the link.</xs:documentation> <xs:appinfo> <uiTip>Comment about the link.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the link."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="download" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A link to a download for the software, e.g. source code, virtual machine image or container.</xs:documentation> <xs:appinfo> <uiTip>A link to a download for the software, e.g. source code, virtual machine image or container.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>Link to download (or repository providing a download) for the software.</xs:documentation> <xs:appinfo> <uiTip>Download link.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="type"> <xs:annotation> <xs:documentation>Type of download that is linked to.</xs:documentation> <xs:appinfo> <uiTip>Download type.</uiTip> <enum> <enumItem> <term>API specification</term> <uiTip>File providing an API specification for the software, e.g. Swagger/OpenAPI, WSDL or RAML file.</uiTip> </enumItem> <enumItem> <term>Biological data</term> <uiTip>Biological data, or a web page on a database portal where such data may be downloaded. </uiTip> </enumItem> <enumItem> <term>Binaries</term> <uiTip>Binaries for the software; compiled code that allow a program to be installed without having to compile the source code.</uiTip> </enumItem> <enumItem> <term>Command-line specification</term> <uiTip>File providing a command line specification for the software.</uiTip> </enumItem> <enumItem> <term>Container file</term> <uiTip>Container file including the software.</uiTip> </enumItem> <enumItem> <term>Icon</term> <uiTip>Icon of the software.</uiTip> </enumItem> <enumItem> <term>Screenshot</term> <uiTip>Screenshot of the software.</uiTip> </enumItem> <enumItem> <term>Source code</term> <uiTip>The source code for the software, that can be compiled or assembled into an executable computer program.</uiTip> </enumItem> <enumItem> <term>Software package</term> <uiTip>A software package; a bundle of files and information about those files, typically including source code and / or binaries.</uiTip> </enumItem> <enumItem> <term>Test data</term> <uiTip>Data for testing the scientific performance of the software or whether it is working correctly.</uiTip> </enumItem> <enumItem> <term>Test script</term> <uiTip>Script used for testing testing whether the software is working correctly.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (CWL)</term> <uiTip>Tool wrapper in Common Workflow Language (CWL) format for the software.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (Galaxy)</term> <uiTip>Galaxy tool configuration file (wrapper) for the software.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (Taverna)</term> <uiTip>Taverna configuration file for the software. </uiTip> </enumItem> <enumItem> <term>Tool wrapper (Other)</term> <uiTip>Workbench configuration file (other than taverna, galaxy or CWL wrapper) for the software.</uiTip> </enumItem> <enumItem> <term>VM image</term> <uiTip>Virtual machine (VM) image for the software.</uiTip> </enumItem> <enumItem> <term>Downloads page</term> <uiTip>Web page summarising general downloads available for the software.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Other type of download for software - the default if a more specific type is not available.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="API specification"/> <xs:enumeration value="Biological data"/> <xs:enumeration value="Binaries"/> <xs:enumeration value="Command-line specification"/> <xs:enumeration value="Container file"/> <xs:enumeration value="Icon"/> <xs:enumeration value="Software package"/> <xs:enumeration value="Screenshot"/> <xs:enumeration value="Source code"/> <xs:enumeration value="Test data"/> <xs:enumeration value="Test script"/> <xs:enumeration value="Tool wrapper (CWL)"/> <xs:enumeration value="Tool wrapper (Galaxy)"/> <xs:enumeration value="Tool wrapper (Taverna)"/> <xs:enumeration value="Tool wrapper (Other)"/> <xs:enumeration value="VM image"/> <xs:enumeration value="Downloads page"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the download.</xs:documentation> <xs:appinfo> <uiTip>Comment about the download.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the download."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="version" type="versionType" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this download.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="documentation" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A link to documentation about the software e.g. manual, API specification or training material.</xs:documentation> <xs:appinfo> <uiTip>A link to documentation about the software e.g. manual, API specification or training material.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="linkType"> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>Link to documentation on the web for the tool.</xs:documentation> <xs:documentation> <uiTip>Documentation link.</uiTip> </xs:documentation> </xs:annotation> </xs:element> <xs:element name="type" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of documentation that is linked to.</xs:documentation> <xs:appinfo> <uiTip>Documentation type.</uiTip> <enum> <enumItem> <term>API documentation</term> <uiTip>Human-readable API documentation.</uiTip> </enumItem> <enumItem> <term>Citation instructions</term> <uiTip>Information on how to correctly cite use of the software; typically which publication(s) to cite, or something more general, e.g. a form of words to use.</uiTip> </enumItem> <enumItem> <term>Code of conduct</term> <uiTip>A set of guidelines or rules outlining the norms, expectations, responsibilities and proper practice for individuals working within the software project.</uiTip> </enumItem> <enumItem> <term>Command-line options</term> <uiTip>Information about the command-line interface to a tool.</uiTip> </enumItem> <enumItem> <term>Contributions policy</term> <uiTip>Information about policy for making contributions to the software project.</uiTip> </enumItem> <enumItem> <term>FAQ</term> <uiTip>Frequently Asked Questions (and answers) about the software.</uiTip> </enumItem> <enumItem> <term>General</term> <uiTip>General documentation.</uiTip> </enumItem> <enumItem> <term>Governance</term> <uiTip>Information about the software governance model.</uiTip> </enumItem> <enumItem> <term>Installation instructions</term> <uiTip>Instructions how to install the software.</uiTip> </enumItem> <enumItem> <term>Quick start guide</term> <uiTip>A short guide helping the end-user to use the software as soon as possible.</uiTip> </enumItem> <enumItem> <term>Release notes</term> <uiTip>Notes about a software release or changes to the software; a change log.</uiTip> </enumItem> <enumItem> <term>Terms of use</term> <uiTip>Rules that one must agree to abide by in order to use a service.</uiTip> </enumItem> <enumItem> <term>Training material</term> <uiTip>Online training material such as text on a Web page, a presentation, video etc. (but excluding tutorials).</uiTip> </enumItem> <enumItem> <term>User manual</term> <uiTip>Information on how to use the software, tailored to the end-user.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Some other type of documentation not listed in biotoolsSchema.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="API documentation"/> <xs:enumeration value="Citation instructions"/> <xs:enumeration value="Code of conduct"/> <xs:enumeration value="Command-line options"/> <xs:enumeration value="Contributions policy"/> <xs:enumeration value="FAQ"/> <xs:enumeration value="General"/> <xs:enumeration value="Governance"/> <xs:enumeration value="Installation instructions"/> <xs:enumeration value="Quick start guide"/> <xs:enumeration value="Release notes"/> <xs:enumeration value="Terms of use"/> <xs:enumeration value="Training material"/> <xs:enumeration value="User manual"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the documentation.</xs:documentation> <xs:appinfo> <uiTip>Comment about the documentation.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the documentation."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="relation" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of a relationship this software shares with other software registered in bio.tools.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="biotoolsID" type="biotoolsIdType"> <xs:annotation> <xs:documentation>bio.tools ID of an existing bio.tools entry which this software is related to.</xs:documentation> <xs:documentation>Relations may only be defined between registered software. The ID is a URL in the bio.tools namespace and reflects (normally exactly) the tool name and version: see http://biotools.readthedocs.io/.</xs:documentation> <xs:appinfo> <uiTip>bio.tools ID (URL) of an existing bio.tools entry which this software is related to.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="type"> <xs:annotation> <xs:documentation>Type of relation between this and another registered software.</xs:documentation> <xs:documentation>Certain relations may only be defined between certain types of tool.</xs:documentation> <xs:appinfo> <uiTip>Type of relation between this and another registered software.</uiTip> <enum> <enumItem> <term>isNewVersionOf / hasNewVersion</term> <uiTip>The software is a new version of an existing software, typically providing new or improved functionality.</uiTip> </enumItem> <enumItem> <term>uses / usedBy</term> <uiTip>The software provides an interface to or in some other way uses the functions of other software under the hood,e.g. invoking a command-line tool or calling a Web API, Web service or SPARQL endpoint to perform its function.</uiTip> </enumItem> <enumItem> <term>includes / includedIn</term> <uiTip>A workbench, toolkit or workflow includes some other, independently available, software.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="isNewVersionOf"/> <xs:enumeration value="hasNewVersion"/> <xs:enumeration value="uses"/> <xs:enumeration value="usedBy"/> <xs:enumeration value="includes"/> <xs:enumeration value="includedIn"/> </xs:restriction> </xs:simpleType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="publication" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A publication about the software.</xs:documentation> <xs:appinfo> <uiTip>A publication about the software.</uiTip> > </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:choice> <xs:sequence> <xs:element name="doi" type="doiType"> <xs:annotation> <xs:appinfo> <uiTip>Digital Object Identifier.</uiTip> > </xs:appinfo> <xs:documentation>Digital Object Identifier (DOI) of a publication about the software.</xs:documentation> </xs:annotation> </xs:element> <xs:element ref="pmid" minOccurs="0"/> <xs:element ref="pmcid" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="pmid"/> <xs:element ref="pmcid" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="pmcid"/> </xs:sequence> </xs:choice> <xs:element name="type" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of publication. </xs:documentation> <xs:appinfo> <uiTip>Type of publication.</uiTip> <enum> <enumItem> <term>Primary</term> <uiTip>The principal publication about the tool itself; the article to cite when acknowledging use of the tool.</uiTip> </enumItem> <enumItem> <term>Method</term> <uiTip>A publication describing a scientific method or algorithm implemented by the tool.</uiTip> </enumItem> <enumItem> <term>Usage</term> <uiTip>A publication describing the application of the tool to scientific research, a particular task or dataset.</uiTip> </enumItem> <enumItem> <term>Benchmarking study</term> <uiTip>A publication which assessed the performance of the tool.</uiTip> </enumItem> <enumItem> <term>Review</term> <uiTip>A publication where the tool was reviewed.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>A publication of relevance to the tool but not fitting the other categories.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Primary"/> <xs:enumeration value="Benchmarking study"/> <xs:enumeration value="Method"/> <xs:enumeration value="Usage"/> <xs:enumeration value="Review"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="version" type="versionType" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this publication.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the publication.</xs:documentation> <xs:appinfo> <uiTip>Comment about the publication.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the publication."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="credit" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>An individual or organisation that should be credited, or may be contacted about the software.</xs:documentation> <xs:appinfo> <uiTip>An individual or organisation that should be credited, or may be contacted about the software.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:choice> <xs:sequence> <xs:element name="name"> <xs:annotation> <xs:appinfo> <uiTip>Name of credited entity.</uiTip> </xs:appinfo> <xs:documentation>Name of the entity that is credited.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Person or institute name"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:minLength value="1"/> <xs:maxLength value="100"/> <xs:whiteSpace value="collapse"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element ref="email" minOccurs="0"/> <xs:element ref="url" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="email"/> <xs:element ref="url" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="url"/> </xs:sequence> </xs:choice> <xs:sequence> <xs:element name="orcidid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>ORCID iD of person.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (ORCID iD) of a person that is credited.</xs:documentation> <xs:documentation>Open Researcher and Contributor IDs (ORCID IDs) provide a persistent reference to information on a researcher, see http://orcid.org/. </xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="https://orcid.org/0000-0002-1825-0097"/> <altova:example value="http://orcid.org/0000-0002-1825-009X"/> <altova:example value="https://orcid.org/0000-0002-1825-009X"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="http://orcid\.org/[0-9]{4}-[0-9]{4}-[0-9]{4}-[0-9]{3}[0-9X]"/> <xs:pattern value="https://orcid\.org/[0-9]{4}-[0-9]{4}-[0-9]{4}-[0-9]{3}[0-9X]"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="gridid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>GRID ID of organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (GRID ID) of an organisation that is credited. </xs:documentation> <xs:documentation>Global Research Identifier Database (GRID) IDs provide a persistent reference to information on research organisations, see https://www.grid.ac/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="grid.5170.3"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="grid.[0-9]{4,}.[a-f0-9]{1,2}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="rorid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>ROR ID of organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (ROR ID) of an organisation that is credited.</xs:documentation> <xs:documentation>Research Organization Registry (ROR) IDs provide a persistent reference to information on research organisations, see https://ror.org/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="03yrm5c26"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="0[0-9a-zA-Z]{6}[0-9]{2}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="fundrefid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>Crossref funder ID of a funding organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (FundRef ID or Funder ID) of a funding organisation that is credited.</xs:documentation> <xs:documentation>The Funder Registry (formerly FundRef) IDs provide a persistent reference to information on funding organisations registered in the Crossref registry, see https://www.crossref.org/services/funder-registry/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.13039/100009273"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="10\.13039/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="typeEntity" minOccurs="0"> <xs:annotation> <xs:documentation>Type of entity that is credited.</xs:documentation> <xs:appinfo> <uiTip>Type of entity that is credited.</uiTip> <enum> <enumItem> <term>Person</term> <uiTip>Credit of an individual.</uiTip> </enumItem> <enumItem> <term>Project</term> <uiTip>Credit of a community software project not formally associated with any single institute.</uiTip> </enumItem> <enumItem> <term>Division</term> <uiTip>Credit of or a formal part of an institutional organisation, e.g. a department, research group, team, etc</uiTip> </enumItem> <enumItem> <term>Institute</term> <uiTip>Credit of an organisation such as a university, hospital, research institute, service center, unit etc.</uiTip> </enumItem> <enumItem> <term>Consortium</term> <uiTip>Credit of an association of two or more institutes or other legal entities which have joined forces for some common purpose. Includes Research Infrastructures (RIs) such as ELIXIR, parts of an RI such as an ELIXIR node etc. </uiTip> </enumItem> <enumItem> <term>Funding agency</term> <uiTip>Credit of a legal entity providing funding for development of the software or provision of an online service.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Person"/> <xs:enumeration value="Project"/> <xs:enumeration value="Division"/> <xs:enumeration value="Institute"/> <xs:enumeration value="Consortium"/> <xs:enumeration value="Funding agency"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="typeRole" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:appinfo> <uiTip>Role performed by credited entity.</uiTip> <enum> <enumItem> <term>Developer</term> <uiTip>Author of the original software source code.</uiTip> </enumItem> <enumItem> <term>Maintainer</term> <uiTip>Maintainer of a mature software providing packaging, patching, distribution etc.</uiTip> </enumItem> <enumItem> <term>Provider</term> <uiTip>Institutional provider of an online service.</uiTip> </enumItem> <enumItem> <term>Documentor</term> <uiTip>Author of software documentation including making edits to a bio.tools entry.</uiTip> </enumItem> <enumItem> <term>Contributor</term> <uiTip>Some other role in software production or service delivery including design, deployment, system administration, evaluation, testing, documentation, training, user support etc.</uiTip> </enumItem> <enumItem> <term>Support</term> <uiTip>Provider of support in using the software.</uiTip> </enumItem> <enumItem> <term>Primary contact</term> <uiTip>The primary point of contact for the software.</uiTip> </enumItem> </enum> </xs:appinfo> <xs:documentation>Role performed by entity that is credited.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Primary contact"/> <xs:enumeration value="Contributor"/> <xs:enumeration value="Developer"/> <xs:enumeration value="Documentor"/> <xs:enumeration value="Maintainer"/> <xs:enumeration value="Provider"/> <xs:enumeration value="Support"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>A comment about the credit.</uiTip> </xs:appinfo> <xs:documentation>A comment about the credit.</xs:documentation> <xs:documentation>This can elaborate on the contribution of the credited entity.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the credit."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element tool/name
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | nameType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="name" type="nameType"> <xs:annotation> <xs:documentation>Canonical software name assigned by the software developer or service provider.</xs:documentation> <xs:appinfo> <uiTip>Software name.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="needle"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/description
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | restriction of textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="description"> <xs:annotation> <xs:documentation>Textual description of the software.</xs:documentation> <xs:documentation>This can be a few sentences copy-pasted from the software homepage.</xs:documentation> <xs:appinfo> <uiTip>Software description, e.g. a few sentences adapted from the software publication abstract or homepage.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="needle reads two input sequences and writes their optimal global sequence alignment to file. It uses the Needleman-Wunsch alignment algorithm to find the optimum alignment (including gaps) of two sequences along their entire length. The algorithm uses a dynamic programming method to ensure the alignment is optimum, by exploring all possible alignments and choosing the best."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="textType"> <xs:maxLength value="1000"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/homepage
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | urlftpType | |||||||||
| properties |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="homepage" type="urlftpType"> <xs:annotation> <xs:documentation>Homepage of the software, or some URL that best serves this purpose.</xs:documentation> <xs:appinfo> <uiTip>Homepage of the software, or some URL that best serves this purpose.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="http://emboss.open-bio.org/rel/rel6/apps/needle.html"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/biotoolsID
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | biotoolsIdType | ||||||
| properties |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="biotoolsID" type="biotoolsIdType" minOccurs="0"> <xs:annotation> <xs:documentation>Unique ID (case insensitive) of the tool that is assigned upon registration of the software in bio.tools, normally identical to tool name.</xs:documentation> <xs:appinfo> <uiTip>Unique ID of the tool that is assigned upon registration of the software in bio.tools.</uiTip> </xs:appinfo> <xs:documentation>bio.tools IDs are set by bio.tools admin and will be disregarded if specified in a payload (e.g. PUSH, POST) to the bio.tools API.</xs:documentation> </xs:annotation> </xs:element> |
element tool/biotoolsCURIE
| diagram |  |
||||||||||
| namespace | biotoolsSchema | ||||||||||
| type | restriction of xs:anyURI | ||||||||||
| properties |
|
||||||||||
| facets |
|
||||||||||
| annotation |
|
||||||||||
| source | <xs:element name="biotoolsCURIE" minOccurs="0"> <xs:annotation> <xs:documentation>bio.tools CURIE (compact URI) based on the bio.tools tool ID.</xs:documentation> <xs:documentation>The bio.tools CURIE is simply the bio.tools tool ID with the prefix "biotools:".</xs:documentation> <xs:appinfo> <uiTip>bio.tools CURIE (compact URI) based on the bio.tools tool ID.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="biotools:signalp"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>bio.tools CURIEs are set by bio.tools admin and will be disregarded if specified in a payload (e.g. PUSH, POST) to the bio.tools API.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="biotools:[_\-.0-9a-zA-Z]*"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/version
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of versionType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="version" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this bio.tools entry.</xs:documentation> <xs:appinfo> <uiTip>Version information (typically a version number) of the software applicable to this bio.tools entry.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="versionType"/> </xs:simpleType> </xs:element> |
element tool/otherID
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| properties |
|
||||||
| children | value type version | ||||||
| annotation |
|
||||||
| source | <xs:element name="otherID" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A unique identifier of the software, typically assigned by an ID-assignment authority other than bio.tools.</xs:documentation> <xs:appinfo> <uiTip>Unique identifier of the software not (typically) assigned by bio.tools.</uiTip> </xs:appinfo> <xs:documentation>This field is not normally used for bio.tools toolIDs, but could be in the exceptional case that multiple such IDs were needed for a given entry.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="value"> <xs:annotation> <xs:documentation>Value of tool identifier.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.1038/nmeth.1701"/> <altova:example value="RRID:SCR_001156"/> <altova:example value="cpe:2.3:a:microsoft:internet_explorer:8.0.6001:beta:*:*:*:*:*:*"/> <altova:example value="biotools:signalp"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>Value of tool identifier.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="10\.[0-9]{4,9}/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> <xs:pattern value="(rrid|RRID):.+"/> <xs:pattern value="(cpe|CPE):.+"/> <xs:pattern value="(BIOTOOLS|biotools):[_\-.0-9a-zA-Z]*"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="type" minOccurs="0"> <xs:annotation> <xs:documentation>Type of tool identifier.</xs:documentation> <xs:appinfo> <uiTip>Type of tool identifier.</uiTip> <enum> <enumItem> <term>doi</term> <uiTip>Digital Object Identifier of the software assigned (typically) by the software developer or service provider.</uiTip> </enumItem> <enumItem> <term>rrid</term> <uiTip>Research Resource Identifier as used by the NIH-supported Resource Identification Portal (https://scicrunch.org/resources).</uiTip> </enumItem> <enumItem> <term>cpe</term> <uiTip>Common Platform Enumeration (CPE) identifier as listed in the CPE dictionary (https://cpe.mitre.org/dictionary/).</uiTip> </enumItem> <enumItem> <term>biotoolsCURIE</term> <uiTip>bio.tools CURIE (secondary identifier).</uiTip> </enumItem> </enum> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="doi"/> <altova:example value="rrid"/> <altova:example value="cpe"/> <altova:example value="biotoolsCURIE"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="doi"/> <xs:enumeration value="rrid"/> <xs:enumeration value="cpe"/> <xs:enumeration value="biotoolsCURIE"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="version" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this identifier.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="versionType"/> </xs:simpleType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element tool/otherID/value
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of xs:token | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="value"> <xs:annotation> <xs:documentation>Value of tool identifier.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.1038/nmeth.1701"/> <altova:example value="RRID:SCR_001156"/> <altova:example value="cpe:2.3:a:microsoft:internet_explorer:8.0.6001:beta:*:*:*:*:*:*"/> <altova:example value="biotools:signalp"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>Value of tool identifier.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="10\.[0-9]{4,9}/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> <xs:pattern value="(rrid|RRID):.+"/> <xs:pattern value="(cpe|CPE):.+"/> <xs:pattern value="(BIOTOOLS|biotools):[_\-.0-9a-zA-Z]*"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/otherID/type
| diagram |  |
||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| annotation |
|
||||||||||||||||||
| source | <xs:element name="type" minOccurs="0"> <xs:annotation> <xs:documentation>Type of tool identifier.</xs:documentation> <xs:appinfo> <uiTip>Type of tool identifier.</uiTip> <enum> <enumItem> <term>doi</term> <uiTip>Digital Object Identifier of the software assigned (typically) by the software developer or service provider.</uiTip> </enumItem> <enumItem> <term>rrid</term> <uiTip>Research Resource Identifier as used by the NIH-supported Resource Identification Portal (https://scicrunch.org/resources).</uiTip> </enumItem> <enumItem> <term>cpe</term> <uiTip>Common Platform Enumeration (CPE) identifier as listed in the CPE dictionary (https://cpe.mitre.org/dictionary/).</uiTip> </enumItem> <enumItem> <term>biotoolsCURIE</term> <uiTip>bio.tools CURIE (secondary identifier).</uiTip> </enumItem> </enum> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="doi"/> <altova:example value="rrid"/> <altova:example value="cpe"/> <altova:example value="biotoolsCURIE"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="doi"/> <xs:enumeration value="rrid"/> <xs:enumeration value="cpe"/> <xs:enumeration value="biotoolsCURIE"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/otherID/version
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of versionType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="version" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this identifier.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="versionType"/> </xs:simpleType> </xs:element> |
element tool/toolType
| diagram |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | |||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="toolType" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A type of application software: a discrete software entity can have more than one type.</xs:documentation> <xs:documentation>bio.tools includes all types of bioinformatics tools: application software with well-defined data processing functions (inputs, outputs and operations). When registering a tool, one or more tool types may be assigned, reflecting the different facets of the software being described.</xs:documentation> <xs:appinfo> <uiTip>Type of application software. A tool may have more than one type.</uiTip> > <enum> <enumItem> <term>Bioinformatics portal</term> <uiTip>A web site providing a platform/portal to multiple resources used for research in a focused area, including biological databases, web applications, training resources and so on.</uiTip> </enumItem> <enumItem> <term>Command-line tool</term> <uiTip>A tool with a text-based (command-line) interface.</uiTip> </enumItem> <enumItem> <term>Database portal</term> <uiTip>A Web site providing a portal to a biological database, typically allowing a user to browse, deposit, search, visualise, analyse or download data.</uiTip> </enumItem> <enumItem> <term>Desktop application</term> <uiTip>A tool with a graphical user interface that runs on your desktop environment, e.g. on a PC or mobile device.</uiTip> </enumItem> <enumItem> <term>Library</term> <uiTip>A collection of components that are used to construct other tools. bio.tools scope includes component libraries performing high-level bioinformatics functions but excludes lower-level programming libraries.</uiTip> </enumItem> <enumItem> <term>Ontology</term> <uiTip>A collection of information about concepts, including terms, synonyms, descriptions etc.</uiTip> </enumItem> <enumItem> <term>Plug-in</term> <uiTip>A software component encapsulating a set of related functions, which are not standalone but depend upon (and typically extend the function of) other software for its use, e.g. a JavaScript widget, plug-in, extension or add-on.</uiTip> </enumItem> <enumItem> <term>Script</term> <uiTip>A tool written for some run-time environment (e.g. other applications or an OS shell) that automates the execution of tasks. Often a small program written in a general-purpose languages (e.g. Perl, Python) or some domain-specific languages (e.g. sed).</uiTip> </enumItem> <enumItem> <term>SPARQL endpoint</term> <uiTip>A service that provides queries over an RDF knowledge base via the SPARQL query language and protocol, and returns results via HTTP.</uiTip> </enumItem> <enumItem> <term>Suite</term> <uiTip>A collection of tools which are bundled together into a convenient toolkit. Such tools typically share related functionality, a common user interface and can exchange data conveniently. This includes collections of stand-alone command-line tools, or Web applications within a common portal.</uiTip> </enumItem> <enumItem> <term>Web application</term> <uiTip>A tool with a graphical user interface that runs in your Web browser.</uiTip> </enumItem> <enumItem> <term>Web API</term> <uiTip>An application programming interface (API) consisting of endpoints to a request-response message system accessible via HTTP. Includes everything from simple data-access URLs to RESTful APIs.</uiTip> </enumItem> <enumItem> <term>Web service</term> <uiTip>An API described in a machine readable form (typically WSDL) providing programmatic access via SOAP over HTTP.</uiTip> </enumItem> <enumItem> <term>Workbench</term> <uiTip>An application or suite with a graphical user interface, providing an integrated environment for data analysis which includes or may be extended with any number of functions or tools. Includes workflow systems, platforms, frameworks etc.</uiTip> </enumItem> <enumItem> <term>Workflow</term> <uiTip>A set of tools which have been composed together into a pipeline of some sort. Such tools are (typically) standalone, but are composed for convenience, for instance for batch execution via some workflow engine or script.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Bioinformatics portal"/> <xs:enumeration value="Command-line tool"/> <xs:enumeration value="Database portal"/> <xs:enumeration value="Desktop application"/> <xs:enumeration value="Library"/> <xs:enumeration value="Ontology"/> <xs:enumeration value="Plug-in"/> <xs:enumeration value="Script"/> <xs:enumeration value="SPARQL endpoint"/> <xs:enumeration value="Suite"/> <xs:enumeration value="Web application"/> <xs:enumeration value="Web API"/> <xs:enumeration value="Web service"/> <xs:enumeration value="Workbench"/> <xs:enumeration value="Workflow"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/topic
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of ontologyConcept | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="topic" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>General scientific domain the software serves or other general category (EDAM Topic).</xs:documentation> <xs:documentation>An EDAM Topic concept URL and / or term are specified, e.g. "Proteomics", http://edamontology.org/topic_0121.</xs:documentation> <xs:appinfo> <uiTip>Scientific topic (EDAM Topic).</uiTip> > </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Topic concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Topic URL.</uiTip> > </xs:appinfo> <xs:documentation>The URL must be in the EDAM Topic namespace, i.e. http://edamontology.org/topic_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/topic_0121"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/topic_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Topic term (preferred label or synonym).</xs:documentation> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <uiTip>EDAM Topic term (preferred label or synonym).</uiTip> > </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Proteomics"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> |
element tool/topic/uri
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:anyURI | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Topic concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Topic URL.</uiTip> > </xs:appinfo> <xs:documentation>The URL must be in the EDAM Topic namespace, i.e. http://edamontology.org/topic_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/topic_0121"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/topic_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/topic/term
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | xs:token | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Topic term (preferred label or synonym).</xs:documentation> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <uiTip>EDAM Topic term (preferred label or synonym).</uiTip> > </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Proteomics"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/topic/term
| diagram |  |
||
| namespace | biotoolsSchema | ||
| type | xs:token | ||
| properties |
|
||
| source | <xs:element name="term" type="xs:token"/> |
element tool/operatingSystem
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of enumType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="operatingSystem" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The operating system supported by a downloadable software.</xs:documentation> <xs:appinfo> <uiTip>Operating system supported by a downloadable software package.</uiTip> <enum> <enumItem> <term>Linux</term> <uiTip>All flavours of Linux/UNIX operating systems.</uiTip> </enumItem> <enumItem> <term>Windows</term> <uiTip>All flavours of Microsoft Windows operating system.</uiTip> </enumItem> <enumItem> <term>Mac</term> <uiTip>All flavours of Apple Macintosh operating systems (primarily Mac OS X).</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Linux"/> <xs:enumeration value="Windows"/> <xs:enumeration value="Mac"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/language
| diagram |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="language" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Name of programming language, e.g. used for the software source code or compatible with an API.</xs:documentation> <xs:appinfo> <uiTip>Source code language.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="ActionScript"/> <xs:enumeration value="Ada"/> <xs:enumeration value="AppleScript"/> <xs:enumeration value="Assembly language"/> <xs:enumeration value="AWK"/> <xs:enumeration value="Bash"/> <xs:enumeration value="C"/> <xs:enumeration value="C#"/> <xs:enumeration value="C++"/> <xs:enumeration value="COBOL"/> <xs:enumeration value="ColdFusion"/> <xs:enumeration value="CWL"/> <xs:enumeration value="D"/> <xs:enumeration value="Delphi"/> <xs:enumeration value="Dylan"/> <xs:enumeration value="Eiffel"/> <xs:enumeration value="Elm"/> <xs:enumeration value="Forth"/> <xs:enumeration value="Fortran"/> <xs:enumeration value="Groovy"/> <xs:enumeration value="Haskell"/> <xs:enumeration value="Icarus"/> <xs:enumeration value="Java"/> <xs:enumeration value="JavaScript"/> <xs:enumeration value="Julia"/> <xs:enumeration value="JSP"/> <xs:enumeration value="LabVIEW"/> <xs:enumeration value="Lisp"/> <xs:enumeration value="Lua"/> <xs:enumeration value="Maple"/> <xs:enumeration value="Mathematica"/> <xs:enumeration value="MATLAB"/> <xs:enumeration value="MLXTRAN"/> <xs:enumeration value="NMTRAN"/> <xs:enumeration value="OCaml"/> <xs:enumeration value="Pascal"/> <xs:enumeration value="Perl"/> <xs:enumeration value="PHP"/> <xs:enumeration value="Prolog"/> <xs:enumeration value="PyMOL"/> <xs:enumeration value="Python"/> <xs:enumeration value="R"/> <xs:enumeration value="Racket"/> <xs:enumeration value="REXX"/> <xs:enumeration value="Ruby"/> <xs:enumeration value="SAS"/> <xs:enumeration value="Scala"/> <xs:enumeration value="Scheme"/> <xs:enumeration value="Shell"/> <xs:enumeration value="Smalltalk"/> <xs:enumeration value="SQL"/> <xs:enumeration value="Turing"/> <xs:enumeration value="Verilog"/> <xs:enumeration value="VHDL"/> <xs:enumeration value="Visual Basic"/> <xs:enumeration value="XAML"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/license
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="license" minOccurs="0"> <xs:annotation> <xs:documentation>Software or data usage license.</xs:documentation> <xs:appinfo> <uiTip>Software or data usage license. For online services, this refers to the license of the underlying software or data.</uiTip> </xs:appinfo> <xs:documentation>Identifier from the SPDX license list (https://spdx.org/licenses/). In future, based on the supplied license, a label e.g. "Open-source" may be applied to the entry in bio.tools.</xs:documentation> <xs:appinfo> <enum> <enumItem> <term>Open-source</term> <uiTip>Software is made available under a license approved by the Open Source Initiative (OSI). The software is distributed in a way that satisfies the 10 criteria of the Open Source Definition maintained by OSI (see https://opensource.org/docs/osd). The source code is available to others.</uiTip> </enumItem> <enumItem> <term>Free software</term> <uiTip>Free as in “freedom” not necessarily free of charge. Software is made available under a license approved by the Free Software Foundation (FSF). The software satisfies the criteria of the Free Software Definition maintained by FSF (see http://www.gnu.org/philosophy/free-sw.html). The source code is available to others.</uiTip> </enumItem> <enumItem> <term>Free and open source</term> <uiTip>Software is made available under a license approved by both the Open Source Initiative (OSI) and the Free Software Foundation (FSF), and satisfies the criteria of the OSI Open Source Definition maintained (https://opensource.org/docs/osd) and the FSF Free Software Definition (http://www.gnu.org/philosophy/free-sw.html). Such software ensures users have the freedom to run, copy, distribute, study, change and improve the software. The source code is available to others.</uiTip> </enumItem> <enumItem> <term>Copyleft</term> <uiTip>Software is made available under a license designated as “copyleft” by the Free Software Foundation (FSF). The license ensures such software is free and that all modified and extended versions of the program are free as well. Free as in “freedom” not necessarily free of charge, as per the Free Software Definition maintained by FSF (see http://www.gnu.org/philosophy/free-sw.html).</uiTip> </enumItem> <enumItem> <term>Proprietary</term> <uiTip>Software for which the software's publisher or another person retains intellectual property rights—usually copyright of the source code, but sometimes patent rights.</uiTip> </enumItem> </enum> <enumItem> <term>Freeware</term> <uiTip>Proprietary software that is available for use at no monetary cost. In other words, freeware may be used without payment but may usually not be modified, re-distributed or reverse-engineered without the author's permission.</uiTip> </enumItem> <enumItem> <term>Not licensed</term> <uiTip>Software which is not licensed and is not proprietary.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Software under license not currently supported by biotoolsSchema.</uiTip> </enumItem> </xs:appinfo> <xs:documentation>Use "Proprietary" where the software must be obtained from the provider (e.g. for money), and is then owned. Use "Freeware" for proprietary software that is available at no monetary cost. Use "Not licensed" for software which is not licensed and is not proprietary, and "Other" for software under license not currently supported by biotoolsSchema.</xs:documentation> <xs:documentation>Note that for online services, the license attribute refers to the license of the underlying software or data, or a part of those.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="0BSD"/> <xs:enumeration value="AAL"/> <xs:enumeration value="ADSL"/> <xs:enumeration value="AFL-1.1"/> <xs:enumeration value="AFL-1.2"/> <xs:enumeration value="AFL-2.0"/> <xs:enumeration value="AFL-2.1"/> <xs:enumeration value="AFL-3.0"/> <xs:enumeration value="AGPL-1.0"/> <xs:enumeration value="AGPL-3.0"/> <xs:enumeration value="AMDPLPA"/> <xs:enumeration value="AML"/> <xs:enumeration value="AMPAS"/> <xs:enumeration value="ANTLR-PD"/> <xs:enumeration value="APAFML"/> <xs:enumeration value="APL-1.0"/> <xs:enumeration value="APSL-1.0"/> <xs:enumeration value="APSL-1.1"/> <xs:enumeration value="APSL-1.2"/> <xs:enumeration value="APSL-2.0"/> <xs:enumeration value="Abstyles"/> <xs:enumeration value="Adobe-2006"/> <xs:enumeration value="Adobe-Glyph"/> <xs:enumeration value="Afmparse"/> <xs:enumeration value="Aladdin"/> <xs:enumeration value="Apache-1.0"/> <xs:enumeration value="Apache-1.1"/> <xs:enumeration value="Apache-2.0"/> <xs:enumeration value="Artistic-1.0"/> <xs:enumeration value="Artistic-1.0-Perl"/> <xs:enumeration value="Artistic-1.0-cl8"/> <xs:enumeration value="Artistic-2.0"/> <xs:enumeration value="BSD-2-Clause"/> <xs:enumeration value="BSD-2-Clause-FreeBSD"/> <xs:enumeration value="BSD-2-Clause-NetBSD"/> <xs:enumeration value="BSD-3-Clause"/> <xs:enumeration value="BSD-3-Clause-Attribution"/> <xs:enumeration value="BSD-3-Clause-Clear"/> <xs:enumeration value="BSD-3-Clause-LBNL"/> <xs:enumeration value="BSD-3-Clause-No-Nuclear-License"/> <xs:enumeration value="BSD-3-Clause-No-Nuclear-License-2014"/> <xs:enumeration value="BSD-3-Clause-No-Nuclear-Warranty"/> <xs:enumeration value="BSD-4-Clause"/> <xs:enumeration value="BSD-4-Clause-UC"/> <xs:enumeration value="BSD-Protection"/> <xs:enumeration value="BSD-Source-Code"/> <xs:enumeration value="BSL-1.0"/> <xs:enumeration value="Bahyph"/> <xs:enumeration value="Barr"/> <xs:enumeration value="Beerware"/> <xs:enumeration value="BitTorrent-1.0"/> <xs:enumeration value="BitTorrent-1.1"/> <xs:enumeration value="Borceux"/> <xs:enumeration value="CATOSL-1.1"/> <xs:enumeration value="CC-BY-1.0"/> <xs:enumeration value="CC-BY-2.0"/> <xs:enumeration value="CC-BY-2.5"/> <xs:enumeration value="CC-BY-3.0"/> <xs:enumeration value="CC-BY-4.0"/> <xs:enumeration value="CC-BY-NC-1.0"/> <xs:enumeration value="CC-BY-NC-2.0"/> <xs:enumeration value="CC-BY-NC-2.5"/> <xs:enumeration value="CC-BY-NC-3.0"/> <xs:enumeration value="CC-BY-NC-4.0"/> <xs:enumeration value="CC-BY-NC-ND-1.0"/> <xs:enumeration value="CC-BY-NC-ND-2.0"/> <xs:enumeration value="CC-BY-NC-ND-2.5"/> <xs:enumeration value="CC-BY-NC-ND-3.0"/> <xs:enumeration value="CC-BY-NC-ND-4.0"/> <xs:enumeration value="CC-BY-NC-SA-1.0"/> <xs:enumeration value="CC-BY-NC-SA-2.0"/> <xs:enumeration value="CC-BY-NC-SA-2.5"/> <xs:enumeration value="CC-BY-NC-SA-3.0"/> <xs:enumeration value="CC-BY-NC-SA-4.0"/> <xs:enumeration value="CC-BY-ND-1.0"/> <xs:enumeration value="CC-BY-ND-2.0"/> <xs:enumeration value="CC-BY-ND-2.5"/> <xs:enumeration value="CC-BY-ND-3.0"/> <xs:enumeration value="CC-BY-ND-4.0"/> <xs:enumeration value="CC-BY-SA-1.0"/> <xs:enumeration value="CC-BY-SA-2.0"/> <xs:enumeration value="CC-BY-SA-2.5"/> <xs:enumeration value="CC-BY-SA-3.0"/> <xs:enumeration value="CC-BY-SA-4.0"/> <xs:enumeration value="CC0-1.0"/> <xs:enumeration value="CDDL-1.0"/> <xs:enumeration value="CDDL-1.1"/> <xs:enumeration value="CECILL-1.0"/> <xs:enumeration value="CECILL-1.1"/> <xs:enumeration value="CECILL-2.0"/> <xs:enumeration value="CECILL-2.1"/> <xs:enumeration value="CECILL-B"/> <xs:enumeration value="CECILL-C"/> <xs:enumeration value="CNRI-Jython"/> <xs:enumeration value="CNRI-Python"/> <xs:enumeration value="CNRI-Python-GPL-Compatible"/> <xs:enumeration value="CPAL-1.0"/> <xs:enumeration value="CPL-1.0"/> <xs:enumeration value="CPOL-1.02"/> <xs:enumeration value="CUA-OPL-1.0"/> <xs:enumeration value="Caldera"/> <xs:enumeration value="ClArtistic"/> <xs:enumeration value="Condor-1.1"/> <xs:enumeration value="Crossword"/> <xs:enumeration value="CrystalStacker"/> <xs:enumeration value="Cube"/> <xs:enumeration value="D-FSL-1.0"/> <xs:enumeration value="DOC"/> <xs:enumeration value="DSDP"/> <xs:enumeration value="Dotseqn"/> <xs:enumeration value="ECL-1.0"/> <xs:enumeration value="ECL-2.0"/> <xs:enumeration value="EFL-1.0"/> <xs:enumeration value="EFL-2.0"/> <xs:enumeration value="EPL-1.0"/> <xs:enumeration value="EUDatagrid"/> <xs:enumeration value="EUPL-1.0"/> <xs:enumeration value="EUPL-1.1"/> <xs:enumeration value="Entessa"/> <xs:enumeration value="ErlPL-1.1"/> <xs:enumeration value="Eurosym"/> <xs:enumeration value="FSFAP"/> <xs:enumeration value="FSFUL"/> <xs:enumeration value="FSFULLR"/> <xs:enumeration value="FTL"/> <xs:enumeration value="Fair"/> <xs:enumeration value="Frameworx-1.0"/> <xs:enumeration value="FreeImage"/> <xs:enumeration value="GFDL-1.1"/> <xs:enumeration value="GFDL-1.2"/> <xs:enumeration value="GFDL-1.3"/> <xs:enumeration value="GL2PS"/> <xs:enumeration value="GPL-1.0"/> <xs:enumeration value="GPL-2.0"/> <xs:enumeration value="GPL-3.0"/> <xs:enumeration value="Giftware"/> <xs:enumeration value="Glide"/> <xs:enumeration value="Glulxe"/> <xs:enumeration value="HPND"/> <xs:enumeration value="HaskellReport"/> <xs:enumeration value="IBM-pibs"/> <xs:enumeration value="ICU"/> <xs:enumeration value="IJG"/> <xs:enumeration value="IPA"/> <xs:enumeration value="IPL-1.0"/> <xs:enumeration value="ISC"/> <xs:enumeration value="ImageMagick"/> <xs:enumeration value="Imlib2"/> <xs:enumeration value="Info-ZIP"/> <xs:enumeration value="Intel"/> <xs:enumeration value="Intel-ACPI"/> <xs:enumeration value="Interbase-1.0"/> <xs:enumeration value="JSON"/> <xs:enumeration value="JasPer-2.0"/> <xs:enumeration value="LAL-1.2"/> <xs:enumeration value="LAL-1.3"/> <xs:enumeration value="LGPL-2.0"/> <xs:enumeration value="LGPL-2.1"/> <xs:enumeration value="LGPL-3.0"/> <xs:enumeration value="LGPLLR"/> <xs:enumeration value="LPL-1.0"/> <xs:enumeration value="LPL-1.02"/> <xs:enumeration value="LPPL-1.0"/> <xs:enumeration value="LPPL-1.1"/> <xs:enumeration value="LPPL-1.2"/> <xs:enumeration value="LPPL-1.3a"/> <xs:enumeration value="LPPL-1.3c"/> <xs:enumeration value="Latex2e"/> <xs:enumeration value="Leptonica"/> <xs:enumeration value="LiLiQ-P-1.1"/> <xs:enumeration value="LiLiQ-R-1.1"/> <xs:enumeration value="LiLiQ-Rplus-1.1"/> <xs:enumeration value="Libpng"/> <xs:enumeration value="MIT"/> <xs:enumeration value="MIT-CMU"/> <xs:enumeration value="MIT-advertising"/> <xs:enumeration value="MIT-enna"/> <xs:enumeration value="MIT-feh"/> <xs:enumeration value="MITNFA"/> <xs:enumeration value="MPL-1.0"/> <xs:enumeration value="MPL-1.1"/> <xs:enumeration value="MPL-2.0"/> <xs:enumeration value="MPL-2.0-no-copyleft-exception"/> <xs:enumeration value="MS-PL"/> <xs:enumeration value="MS-RL"/> <xs:enumeration value="MTLL"/> <xs:enumeration value="MakeIndex"/> <xs:enumeration value="MirOS"/> <xs:enumeration value="Motosoto"/> <xs:enumeration value="Multics"/> <xs:enumeration value="Mup"/> <xs:enumeration value="NASA-1.3"/> <xs:enumeration value="NBPL-1.0"/> <xs:enumeration value="NCSA"/> <xs:enumeration value="NGPL"/> <xs:enumeration value="NLOD-1.0"/> <xs:enumeration value="NLPL"/> <xs:enumeration value="NOSL"/> <xs:enumeration value="NPL-1.0"/> <xs:enumeration value="NPL-1.1"/> <xs:enumeration value="NPOSL-3.0"/> <xs:enumeration value="NRL"/> <xs:enumeration value="NTP"/> <xs:enumeration value="Naumen"/> <xs:enumeration value="NetCDF"/> <xs:enumeration value="Newsletr"/> <xs:enumeration value="Nokia"/> <xs:enumeration value="Noweb"/> <xs:enumeration value="Nunit"/> <xs:enumeration value="OCCT-PL"/> <xs:enumeration value="OCLC-2.0"/> <xs:enumeration value="ODbL-1.0"/> <xs:enumeration value="OFL-1.0"/> <xs:enumeration value="OFL-1.1"/> <xs:enumeration value="OGTSL"/> <xs:enumeration value="OLDAP-1.1"/> <xs:enumeration value="OLDAP-1.2"/> <xs:enumeration value="OLDAP-1.3"/> <xs:enumeration value="OLDAP-1.4"/> <xs:enumeration value="OLDAP-2.0"/> <xs:enumeration value="OLDAP-2.0.1"/> <xs:enumeration value="OLDAP-2.1"/> <xs:enumeration value="OLDAP-2.2"/> <xs:enumeration value="OLDAP-2.2.1"/> <xs:enumeration value="OLDAP-2.2.2"/> <xs:enumeration value="OLDAP-2.3"/> <xs:enumeration value="OLDAP-2.4"/> <xs:enumeration value="OLDAP-2.5"/> <xs:enumeration value="OLDAP-2.6"/> <xs:enumeration value="OLDAP-2.7"/> <xs:enumeration value="OLDAP-2.8"/> <xs:enumeration value="OML"/> <xs:enumeration value="OPL-1.0"/> <xs:enumeration value="OSET-PL-2.1"/> <xs:enumeration value="OSL-1.0"/> <xs:enumeration value="OSL-1.1"/> <xs:enumeration value="OSL-2.0"/> <xs:enumeration value="OSL-2.1"/> <xs:enumeration value="OSL-3.0"/> <xs:enumeration value="OpenSSL"/> <xs:enumeration value="PDDL-1.0"/> <xs:enumeration value="PHP-3.0"/> <xs:enumeration value="PHP-3.01"/> <xs:enumeration value="Plexus"/> <xs:enumeration value="PostgreSQL"/> <xs:enumeration value="Python-2.0"/> <xs:enumeration value="QPL-1.0"/> <xs:enumeration value="Qhull"/> <xs:enumeration value="RHeCos-1.1"/> <xs:enumeration value="RPL-1.1"/> <xs:enumeration value="RPL-1.5"/> <xs:enumeration value="RPSL-1.0"/> <xs:enumeration value="RSA-MD"/> <xs:enumeration value="RSCPL"/> <xs:enumeration value="Rdisc"/> <xs:enumeration value="Ruby"/> <xs:enumeration value="SAX-PD"/> <xs:enumeration value="SCEA"/> <xs:enumeration value="SGI-B-1.0"/> <xs:enumeration value="SGI-B-1.1"/> <xs:enumeration value="SGI-B-2.0"/> <xs:enumeration value="SISSL"/> <xs:enumeration value="SISSL-1.2"/> <xs:enumeration value="SMLNJ"/> <xs:enumeration value="SMPPL"/> <xs:enumeration value="SNIA"/> <xs:enumeration value="SPL-1.0"/> <xs:enumeration value="SWL"/> <xs:enumeration value="Saxpath"/> <xs:enumeration value="Sendmail"/> <xs:enumeration value="SimPL-2.0"/> <xs:enumeration value="Sleepycat"/> <xs:enumeration value="Spencer-86"/> <xs:enumeration value="Spencer-94"/> <xs:enumeration value="Spencer-99"/> <xs:enumeration value="SugarCRM-1.1.3"/> <xs:enumeration value="TCL"/> <xs:enumeration value="TMate"/> <xs:enumeration value="TORQUE-1.1"/> <xs:enumeration value="TOSL"/> <xs:enumeration value="UPL-1.0"/> <xs:enumeration value="Unicode-TOU"/> <xs:enumeration value="Unlicense"/> <xs:enumeration value="VOSTROM"/> <xs:enumeration value="VSL-1.0"/> <xs:enumeration value="Vim"/> <xs:enumeration value="W3C"/> <xs:enumeration value="W3C-19980720"/> <xs:enumeration value="WTFPL"/> <xs:enumeration value="Watcom-1.0"/> <xs:enumeration value="Wsuipa"/> <xs:enumeration value="X11"/> <xs:enumeration value="XFree86-1.1"/> <xs:enumeration value="XSkat"/> <xs:enumeration value="Xerox"/> <xs:enumeration value="Xnet"/> <xs:enumeration value="YPL-1.0"/> <xs:enumeration value="YPL-1.1"/> <xs:enumeration value="ZPL-1.1"/> <xs:enumeration value="ZPL-2.0"/> <xs:enumeration value="ZPL-2.1"/> <xs:enumeration value="Zed"/> <xs:enumeration value="Zend-2.0"/> <xs:enumeration value="Zimbra-1.3"/> <xs:enumeration value="Zimbra-1.4"/> <xs:enumeration value="Zlib"/> <xs:enumeration value="bzip2-1.0.5"/> <xs:enumeration value="bzip2-1.0.6"/> <xs:enumeration value="curl"/> <xs:enumeration value="diffmark"/> <xs:enumeration value="dvipdfm"/> <xs:enumeration value="eGenix"/> <xs:enumeration value="gSOAP-1.3b"/> <xs:enumeration value="gnuplot"/> <xs:enumeration value="iMatix"/> <xs:enumeration value="libtiff"/> <xs:enumeration value="mpich2"/> <xs:enumeration value="psfrag"/> <xs:enumeration value="psutils"/> <xs:enumeration value="xinetd"/> <xs:enumeration value="xpp"/> <xs:enumeration value="zlib-acknowledgement"/> <xs:enumeration value="Proprietary"/> <xs:enumeration value="Other"/> <xs:enumeration value="Not licensed"/> <xs:enumeration value="Freeware"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/collectionID
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of nameType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="collectionID" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A tag which assigns the software to a collection in bio.tools.</xs:documentation> <xs:appinfo> <uiTip>Tag for a collection that the software has been assigned to within bio.tools.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="emboss"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="nameType"/> </xs:simpleType> </xs:element> |
element tool/maturity
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of enumType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="maturity" minOccurs="0"> <xs:annotation> <xs:documentation>Development stage of the software product.</xs:documentation> <xs:appinfo> <uiTip>How mature the software product is.</uiTip> <enum> <enumItem> <term>Emerging</term> <uiTip>Nascent or early release software that may not yet be fully featured or stable.</uiTip> </enumItem> <enumItem> <term>Mature</term> <uiTip>Software that is generally considered to fulfill several of the following: secure, reliable, actively maintained, fully featured, proven in production environments, has an active community, and is described or cited in the scientific literature.</uiTip> </enumItem> <enumItem> <term>Legacy</term> <uiTip>Software which is no longer in common use, deprecated by the provider, superseded by other software, replaced by a newer version, is obsolete etc.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Emerging"/> <xs:enumeration value="Mature"/> <xs:enumeration value="Legacy"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/cost
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of enumType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="cost" minOccurs="0"> <xs:annotation> <xs:documentation>Monetary cost of acquiring the software.</xs:documentation> <xs:appinfo> <uiTip>Monetary cost of acquiring the software.</uiTip> <enum> <enumItem> <term>Free of charge</term> <uiTip>Software which available for use by all, with full functionality, at no monetary cost to the user.</uiTip> </enumItem> <enumItem> <term>Free of charge (with restrictions)</term> <uiTip>Software which is available for use at no monetary cost to the user, but possibly with limited functionality, usage restrictions, or other limitations.</uiTip> </enumItem> <enumItem> <term>Commercial</term> <uiTip>Software which you have to pay to access.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Free of charge"/> <xs:enumeration value="Free of charge (with restrictions)"/> <xs:enumeration value="Commercial"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/accessibility
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | restriction of enumType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="accessibility" minOccurs="0"> <xs:annotation> <xs:documentation>Whether an online service is freely available for use.</xs:documentation> <xs:appinfo> <uiTip>Whether the software is freely available for use.</uiTip> <enum> <enumItem> <term>Open access</term> <uiTip>An online service which is available for use to all, but possibly requiring user accounts / authentication.</uiTip> </enumItem> <enumItem> <term>Restricted access</term> <uiTip>An online service which is available for use to a restricted audience, e.g. members of a specific institute.</uiTip> </enumItem> <enumItem> <term>Proprietary</term> <uiTip>Software for which the software's publisher or another person retains intellectual property rights—usually copyright of the source code, but sometimes patent rights.</uiTip> </enumItem> <enumItem> <term>Freeware</term> <uiTip>Proprietary software that is available for use at no monetary cost. In other words, freeware may be used without payment but may usually not be modified, re-distributed or reverse-engineered without the author's permission.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Open access"/> <xs:enumeration value="Restricted access"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/elixirPlatform
| diagram |  |
|||||||||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||||||||
| type | restriction of enumType | |||||||||||||||||||||
| properties |
|
|||||||||||||||||||||
| facets |
|
|||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||
| source | <xs:element name="elixirPlatform" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>ELIXIR platform credited for developing or providing the software.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Training"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>ELIXIR platform credited for developing or providing the software.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Data"/> <xs:enumeration value="Tools"/> <xs:enumeration value="Compute"/> <xs:enumeration value="Interoperability"/> <xs:enumeration value="Training"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/elixirCommunity
| diagram |  |
|||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | |||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="elixirCommunity" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>ELIXIR (or other) community to which the software is relevant.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="3D-BioInfo"/> <xs:enumeration value="Federated Human Data"/> <xs:enumeration value="Galaxy"/> <xs:enumeration value="Human Copy Number Variation"/> <xs:enumeration value="Intrinsically Disordered Proteins"/> <xs:enumeration value="Marine Metagenomics"/> <xs:enumeration value="Metabolomics"/> <xs:enumeration value="Microbial Biotechnology"/> <xs:enumeration value="Plant Sciences"/> <xs:enumeration value="Proteomics"/> <xs:enumeration value="Rare Diseases"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/elixirNode
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="elixirNode" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>ELIXIR node credited for developing or providing the software - the software is in Node Service Delivery Plan.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="UK"/> </altova:exampleValues> </xs:appinfo> <xs:appinfo> <uiTip>ELIXIR node credited for developing or providing the software - the software is in Node Service Delivery Plan.</uiTip> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Belgium"/> <xs:enumeration value="Czech Republic"/> <xs:enumeration value="Denmark"/> <xs:enumeration value="EMBL"/> <xs:enumeration value="Estonia"/> <xs:enumeration value="Finland"/> <xs:enumeration value="France"/> <xs:enumeration value="Germany"/> <xs:enumeration value="Greece"/> <xs:enumeration value="Hungary"/> <xs:enumeration value="Ireland"/> <xs:enumeration value="Israel"/> <xs:enumeration value="Italy"/> <xs:enumeration value="Luxembourg"/> <xs:enumeration value="Netherlands"/> <xs:enumeration value="Norway"/> <xs:enumeration value="Portugal"/> <xs:enumeration value="Slovenia"/> <xs:enumeration value="Spain"/> <xs:enumeration value="Sweden"/> <xs:enumeration value="Switzerland"/> <xs:enumeration value="UK"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/function
| diagram | 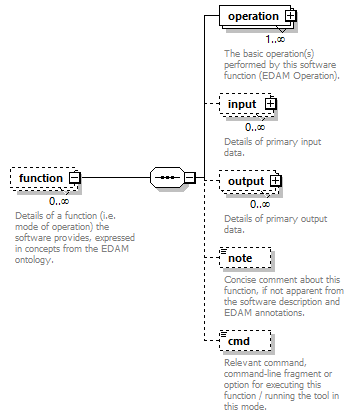 |
||||||
| namespace | biotoolsSchema | ||||||
| properties |
|
||||||
| children | operation input output note cmd | ||||||
| annotation |
|
||||||
| source | <xs:element name="function" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of a function (i.e. mode of operation) the software provides, expressed in concepts from the EDAM ontology.</xs:documentation> <xs:appinfo> <uiTip>Details of a function the software provides, expressed in terms from the EDAM ontology.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="operation" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The basic operation(s) performed by this software function (EDAM Operation).</xs:documentation> <xs:documentation>An EDAM Operation concept URL and / or term are specified, e.g. "Multiple sequence alignment", http://edamontology.org/operation_0492.</xs:documentation> <xs:appinfo> <uiTip>Operation performed (EDAM operation).</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Operation concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation URL.</uiTip> </xs:appinfo> <xs:documentation>The URL must be in the EDAM Operation namespace, i.e. http://edamontology.org/operation_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/operation_0492"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/operation_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Operation term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation term (preferred label or synonym).</uiTip> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Multiple sequence alignment"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="input" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of primary input data.</xs:documentation> <xs:appinfo> <uiTip>Details of primary input data.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="dataType"> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary input data, if any (EDAM data). </xs:documentation> <xs:appinfo> <uiTip>Input data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and / or term are specified, e.g. "Protein sequences", http://edamontology.org/data_2976. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the input data (EDAM Format). </xs:documentation> <xs:appinfo> <uiTip>Input data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and / or term are specified, e.g. "FASTA", http://edamontology.org/format_1929.</xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="output" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of primary output data.</xs:documentation> <xs:appinfo> <uiTip>Details of primary output data.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="dataType"> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary output data, if any (EDAM Data).</xs:documentation> <xs:appinfo> <uiTip>Output data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and (optionally) term are specified, e.g. "Sequence alignment", http://edamontology.org/data_0863.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the output data (EDAM Format).</xs:documentation> <xs:appinfo> <uiTip>Output data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and (optionally) term are specified, e.g. “FASTA-aln”, http://edamontology.org/format_1984.</xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="note" minOccurs="0"> <xs:annotation> <xs:documentation>Concise comment about this function, if not apparent from the software description and EDAM annotations.</xs:documentation> <xs:appinfo> <uiTip>General comment about function.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Concise comment about this function, if not apparent from the software description and EDAM annotations."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="textType"/> </xs:simpleType> </xs:element> <xs:element name="cmd" minOccurs="0"> <xs:annotation> <xs:documentation>Relevant command, command-line fragment or option for executing this function / running the tool in this mode.</xs:documentation> <xs:appinfo> <uiTip>Relevant command, command-line fragment or option for executing this function / running the tool in this mode.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="-s best"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:maxLength value="1000"/> <xs:whiteSpace value="collapse"/> <xs:minLength value="1"/> </xs:restriction> </xs:simpleType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element tool/function/operation
| diagram | 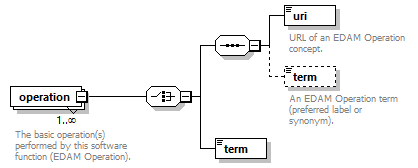 |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of ontologyConcept | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="operation" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The basic operation(s) performed by this software function (EDAM Operation).</xs:documentation> <xs:documentation>An EDAM Operation concept URL and / or term are specified, e.g. "Multiple sequence alignment", http://edamontology.org/operation_0492.</xs:documentation> <xs:appinfo> <uiTip>Operation performed (EDAM operation).</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Operation concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation URL.</uiTip> </xs:appinfo> <xs:documentation>The URL must be in the EDAM Operation namespace, i.e. http://edamontology.org/operation_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/operation_0492"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/operation_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Operation term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation term (preferred label or synonym).</uiTip> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Multiple sequence alignment"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> |
element tool/function/operation/uri
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:anyURI | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Operation concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation URL.</uiTip> </xs:appinfo> <xs:documentation>The URL must be in the EDAM Operation namespace, i.e. http://edamontology.org/operation_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/operation_0492"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/operation_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/function/operation/term
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | xs:token | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Operation term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Operation term (preferred label or synonym).</uiTip> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Multiple sequence alignment"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/function/operation/term
| diagram |  |
||
| namespace | biotoolsSchema | ||
| type | xs:token | ||
| properties |
|
||
| source | <xs:element name="term" type="xs:token"/> |
element tool/function/input
| diagram | 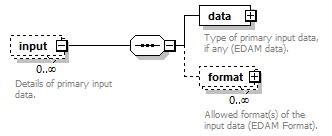 |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of dataType | ||||||
| properties |
|
||||||
| children | data format | ||||||
| annotation |
|
||||||
| source | <xs:element name="input" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of primary input data.</xs:documentation> <xs:appinfo> <uiTip>Details of primary input data.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="dataType"> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary input data, if any (EDAM data). </xs:documentation> <xs:appinfo> <uiTip>Input data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and / or term are specified, e.g. "Protein sequences", http://edamontology.org/data_2976. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the input data (EDAM Format). </xs:documentation> <xs:appinfo> <uiTip>Input data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and / or term are specified, e.g. "FASTA", http://edamontology.org/format_1929.</xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> |
element tool/function/input/data
| diagram | 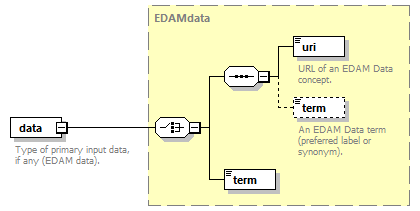 |
||||||
| namespace | biotoolsSchema | ||||||
| type | EDAMdata | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary input data, if any (EDAM data). </xs:documentation> <xs:appinfo> <uiTip>Input data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and / or term are specified, e.g. "Protein sequences", http://edamontology.org/data_2976. </xs:documentation> </xs:annotation> </xs:element> |
element tool/function/input/format
| diagram | 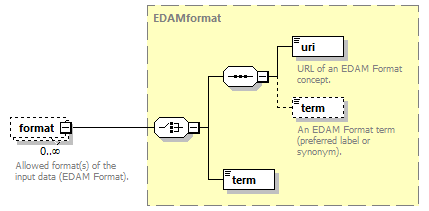 |
||||||
| namespace | biotoolsSchema | ||||||
| type | EDAMformat | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the input data (EDAM Format). </xs:documentation> <xs:appinfo> <uiTip>Input data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and / or term are specified, e.g. "FASTA", http://edamontology.org/format_1929.</xs:appinfo> </xs:annotation> </xs:element> |
element tool/function/output
| diagram | 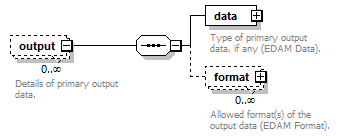 |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of dataType | ||||||
| properties |
|
||||||
| children | data format | ||||||
| annotation |
|
||||||
| source | <xs:element name="output" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of primary output data.</xs:documentation> <xs:appinfo> <uiTip>Details of primary output data.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="dataType"> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary output data, if any (EDAM Data).</xs:documentation> <xs:appinfo> <uiTip>Output data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and (optionally) term are specified, e.g. "Sequence alignment", http://edamontology.org/data_0863.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the output data (EDAM Format).</xs:documentation> <xs:appinfo> <uiTip>Output data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and (optionally) term are specified, e.g. “FASTA-aln”, http://edamontology.org/format_1984.</xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> |
element tool/function/output/data
| diagram | 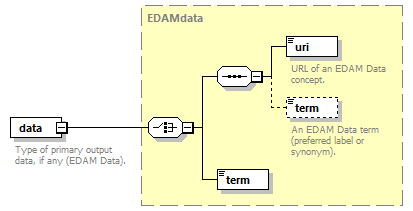 |
||||||
| namespace | biotoolsSchema | ||||||
| type | EDAMdata | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of primary output data, if any (EDAM Data).</xs:documentation> <xs:appinfo> <uiTip>Output data type (EDAM data).</uiTip> </xs:appinfo> <xs:documentation>An EDAM Data concept URL and (optionally) term are specified, e.g. "Sequence alignment", http://edamontology.org/data_0863.</xs:documentation> </xs:annotation> </xs:element> |
element tool/function/output/format
| diagram | 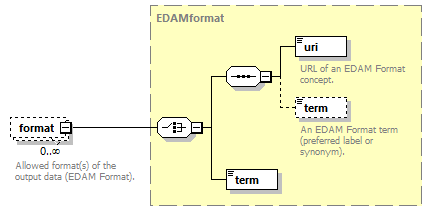 |
||||||
| namespace | biotoolsSchema | ||||||
| type | EDAMformat | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the output data (EDAM Format).</xs:documentation> <xs:appinfo> <uiTip>Output data format (EDAM format).</uiTip> </xs:appinfo> <xs:appinfo>An EDAM Format concept URL and (optionally) term are specified, e.g. “FASTA-aln”, http://edamontology.org/format_1984.</xs:appinfo> </xs:annotation> </xs:element> |
element tool/function/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | restriction of textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" minOccurs="0"> <xs:annotation> <xs:documentation>Concise comment about this function, if not apparent from the software description and EDAM annotations.</xs:documentation> <xs:appinfo> <uiTip>General comment about function.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Concise comment about this function, if not apparent from the software description and EDAM annotations."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="textType"/> </xs:simpleType> </xs:element> |
element tool/function/cmd
| diagram | 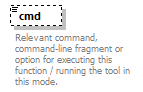 |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | restriction of xs:token | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="cmd" minOccurs="0"> <xs:annotation> <xs:documentation>Relevant command, command-line fragment or option for executing this function / running the tool in this mode.</xs:documentation> <xs:appinfo> <uiTip>Relevant command, command-line fragment or option for executing this function / running the tool in this mode.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="-s best"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:maxLength value="1000"/> <xs:whiteSpace value="collapse"/> <xs:minLength value="1"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/link
| diagram | 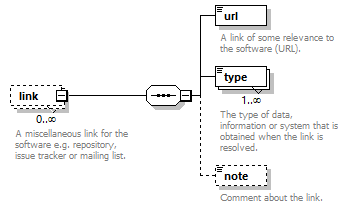 |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of linkType | ||||||
| properties |
|
||||||
| children | url type note | ||||||
| annotation |
|
||||||
| source | <xs:element name="link" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A miscellaneous link for the software e.g. repository, issue tracker or mailing list.</xs:documentation> <xs:appinfo> <uiTip>A miscellaneous link for the software e.g. repository, issue tracker or mailing list.</uiTip> > </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="linkType"> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>A link of some relevance to the software (URL).</xs:documentation> <xs:documentation> <uiTip>Miscellaneous link.</uiTip> </xs:documentation> </xs:annotation> </xs:element> <xs:element name="type" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The type of data, information or system that is obtained when the link is resolved.</xs:documentation> <xs:appinfo> <uiTip>Type of thing linked to.</uiTip> <enum> <enumItem> <term>Discussion forum</term> <uiTip>Online forum for user discussions about the software.</uiTip> </enumItem> <enumItem> <term>Galaxy service</term> <uiTip>An online service providing the tool through the Galaxy platform.</uiTip> </enumItem> <enumItem> <term>Helpdesk</term> <uiTip>A phone line, web site or email-based system providing help to the end-user of the software.</uiTip> </enumItem> <enumItem> <term>Issue tracker</term> <uiTip>Tracker for software issues, bug reports, feature requests etc.</uiTip> </enumItem> <enumItem> <term>Mailing list</term> <uiTip>Mailing list for the software announcements, discussions, support etc.</uiTip> </enumItem> <enumItem> <term>Mirror</term> <uiTip>Mirror of an (identical) online service.</uiTip> </enumItem> <enumItem> <term>Software catalogue</term> <uiTip>Some registry, catalogue etc. other than bio.tools where the tool is also described..</uiTip> </enumItem> <enumItem> <term>Repository</term> <uiTip>A place where source code, data and other files can be retrieved from, typically via platforms like GitHub which provide version control and other features, or something simpler, e.g. an FTP site.</uiTip> </enumItem> <enumItem> <term>Social media</term> <uiTip>A website used by the software community including social networking sites, discussion and support fora, WIKIs etc.</uiTip> </enumItem> <enumItem> <term>Service</term> <uiTip>An online service (other than Galaxy) that provides access (an interface) to the software.</uiTip> </enumItem> <enumItem> <term>Technical monitoring</term> <uiTip>Information about the technical status of a tool.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Other type of link for software - the default if a more specific type is not available.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Discussion forum"/> <xs:enumeration value="Galaxy service"/> <xs:enumeration value="Helpdesk"/> <xs:enumeration value="Issue tracker"/> <xs:enumeration value="Mailing list"/> <xs:enumeration value="Mirror"/> <xs:enumeration value="Software catalogue"/> <xs:enumeration value="Repository"/> <xs:enumeration value="Service"/> <xs:enumeration value="Social media"/> <xs:enumeration value="Technical monitoring"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the link.</xs:documentation> <xs:appinfo> <uiTip>Comment about the link.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the link."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> |
element tool/link/url
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | urlftpType | |||||||||
| properties |
|
|||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>A link of some relevance to the software (URL).</xs:documentation> <xs:documentation> <uiTip>Miscellaneous link.</uiTip> </xs:documentation> </xs:annotation> </xs:element> |
element tool/link/type
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="type" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The type of data, information or system that is obtained when the link is resolved.</xs:documentation> <xs:appinfo> <uiTip>Type of thing linked to.</uiTip> <enum> <enumItem> <term>Discussion forum</term> <uiTip>Online forum for user discussions about the software.</uiTip> </enumItem> <enumItem> <term>Galaxy service</term> <uiTip>An online service providing the tool through the Galaxy platform.</uiTip> </enumItem> <enumItem> <term>Helpdesk</term> <uiTip>A phone line, web site or email-based system providing help to the end-user of the software.</uiTip> </enumItem> <enumItem> <term>Issue tracker</term> <uiTip>Tracker for software issues, bug reports, feature requests etc.</uiTip> </enumItem> <enumItem> <term>Mailing list</term> <uiTip>Mailing list for the software announcements, discussions, support etc.</uiTip> </enumItem> <enumItem> <term>Mirror</term> <uiTip>Mirror of an (identical) online service.</uiTip> </enumItem> <enumItem> <term>Software catalogue</term> <uiTip>Some registry, catalogue etc. other than bio.tools where the tool is also described..</uiTip> </enumItem> <enumItem> <term>Repository</term> <uiTip>A place where source code, data and other files can be retrieved from, typically via platforms like GitHub which provide version control and other features, or something simpler, e.g. an FTP site.</uiTip> </enumItem> <enumItem> <term>Social media</term> <uiTip>A website used by the software community including social networking sites, discussion and support fora, WIKIs etc.</uiTip> </enumItem> <enumItem> <term>Service</term> <uiTip>An online service (other than Galaxy) that provides access (an interface) to the software.</uiTip> </enumItem> <enumItem> <term>Technical monitoring</term> <uiTip>Information about the technical status of a tool.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Other type of link for software - the default if a more specific type is not available.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Discussion forum"/> <xs:enumeration value="Galaxy service"/> <xs:enumeration value="Helpdesk"/> <xs:enumeration value="Issue tracker"/> <xs:enumeration value="Mailing list"/> <xs:enumeration value="Mirror"/> <xs:enumeration value="Software catalogue"/> <xs:enumeration value="Repository"/> <xs:enumeration value="Service"/> <xs:enumeration value="Social media"/> <xs:enumeration value="Technical monitoring"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/link/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the link.</xs:documentation> <xs:appinfo> <uiTip>Comment about the link.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the link."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/download
| diagram | 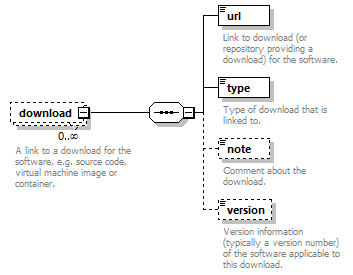 |
||||||
| namespace | biotoolsSchema | ||||||
| properties |
|
||||||
| children | url type note version | ||||||
| annotation |
|
||||||
| source | <xs:element name="download" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A link to a download for the software, e.g. source code, virtual machine image or container.</xs:documentation> <xs:appinfo> <uiTip>A link to a download for the software, e.g. source code, virtual machine image or container.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>Link to download (or repository providing a download) for the software.</xs:documentation> <xs:appinfo> <uiTip>Download link.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="type"> <xs:annotation> <xs:documentation>Type of download that is linked to.</xs:documentation> <xs:appinfo> <uiTip>Download type.</uiTip> <enum> <enumItem> <term>API specification</term> <uiTip>File providing an API specification for the software, e.g. Swagger/OpenAPI, WSDL or RAML file.</uiTip> </enumItem> <enumItem> <term>Biological data</term> <uiTip>Biological data, or a web page on a database portal where such data may be downloaded. </uiTip> </enumItem> <enumItem> <term>Binaries</term> <uiTip>Binaries for the software; compiled code that allow a program to be installed without having to compile the source code.</uiTip> </enumItem> <enumItem> <term>Command-line specification</term> <uiTip>File providing a command line specification for the software.</uiTip> </enumItem> <enumItem> <term>Container file</term> <uiTip>Container file including the software.</uiTip> </enumItem> <enumItem> <term>Icon</term> <uiTip>Icon of the software.</uiTip> </enumItem> <enumItem> <term>Screenshot</term> <uiTip>Screenshot of the software.</uiTip> </enumItem> <enumItem> <term>Source code</term> <uiTip>The source code for the software, that can be compiled or assembled into an executable computer program.</uiTip> </enumItem> <enumItem> <term>Software package</term> <uiTip>A software package; a bundle of files and information about those files, typically including source code and / or binaries.</uiTip> </enumItem> <enumItem> <term>Test data</term> <uiTip>Data for testing the scientific performance of the software or whether it is working correctly.</uiTip> </enumItem> <enumItem> <term>Test script</term> <uiTip>Script used for testing testing whether the software is working correctly.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (CWL)</term> <uiTip>Tool wrapper in Common Workflow Language (CWL) format for the software.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (Galaxy)</term> <uiTip>Galaxy tool configuration file (wrapper) for the software.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (Taverna)</term> <uiTip>Taverna configuration file for the software. </uiTip> </enumItem> <enumItem> <term>Tool wrapper (Other)</term> <uiTip>Workbench configuration file (other than taverna, galaxy or CWL wrapper) for the software.</uiTip> </enumItem> <enumItem> <term>VM image</term> <uiTip>Virtual machine (VM) image for the software.</uiTip> </enumItem> <enumItem> <term>Downloads page</term> <uiTip>Web page summarising general downloads available for the software.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Other type of download for software - the default if a more specific type is not available.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="API specification"/> <xs:enumeration value="Biological data"/> <xs:enumeration value="Binaries"/> <xs:enumeration value="Command-line specification"/> <xs:enumeration value="Container file"/> <xs:enumeration value="Icon"/> <xs:enumeration value="Software package"/> <xs:enumeration value="Screenshot"/> <xs:enumeration value="Source code"/> <xs:enumeration value="Test data"/> <xs:enumeration value="Test script"/> <xs:enumeration value="Tool wrapper (CWL)"/> <xs:enumeration value="Tool wrapper (Galaxy)"/> <xs:enumeration value="Tool wrapper (Taverna)"/> <xs:enumeration value="Tool wrapper (Other)"/> <xs:enumeration value="VM image"/> <xs:enumeration value="Downloads page"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the download.</xs:documentation> <xs:appinfo> <uiTip>Comment about the download.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the download."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="version" type="versionType" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this download.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element tool/download/url
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | urlftpType | |||||||||
| properties |
|
|||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>Link to download (or repository providing a download) for the software.</xs:documentation> <xs:appinfo> <uiTip>Download link.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/download/type
| diagram |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="type"> <xs:annotation> <xs:documentation>Type of download that is linked to.</xs:documentation> <xs:appinfo> <uiTip>Download type.</uiTip> <enum> <enumItem> <term>API specification</term> <uiTip>File providing an API specification for the software, e.g. Swagger/OpenAPI, WSDL or RAML file.</uiTip> </enumItem> <enumItem> <term>Biological data</term> <uiTip>Biological data, or a web page on a database portal where such data may be downloaded. </uiTip> </enumItem> <enumItem> <term>Binaries</term> <uiTip>Binaries for the software; compiled code that allow a program to be installed without having to compile the source code.</uiTip> </enumItem> <enumItem> <term>Command-line specification</term> <uiTip>File providing a command line specification for the software.</uiTip> </enumItem> <enumItem> <term>Container file</term> <uiTip>Container file including the software.</uiTip> </enumItem> <enumItem> <term>Icon</term> <uiTip>Icon of the software.</uiTip> </enumItem> <enumItem> <term>Screenshot</term> <uiTip>Screenshot of the software.</uiTip> </enumItem> <enumItem> <term>Source code</term> <uiTip>The source code for the software, that can be compiled or assembled into an executable computer program.</uiTip> </enumItem> <enumItem> <term>Software package</term> <uiTip>A software package; a bundle of files and information about those files, typically including source code and / or binaries.</uiTip> </enumItem> <enumItem> <term>Test data</term> <uiTip>Data for testing the scientific performance of the software or whether it is working correctly.</uiTip> </enumItem> <enumItem> <term>Test script</term> <uiTip>Script used for testing testing whether the software is working correctly.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (CWL)</term> <uiTip>Tool wrapper in Common Workflow Language (CWL) format for the software.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (Galaxy)</term> <uiTip>Galaxy tool configuration file (wrapper) for the software.</uiTip> </enumItem> <enumItem> <term>Tool wrapper (Taverna)</term> <uiTip>Taverna configuration file for the software. </uiTip> </enumItem> <enumItem> <term>Tool wrapper (Other)</term> <uiTip>Workbench configuration file (other than taverna, galaxy or CWL wrapper) for the software.</uiTip> </enumItem> <enumItem> <term>VM image</term> <uiTip>Virtual machine (VM) image for the software.</uiTip> </enumItem> <enumItem> <term>Downloads page</term> <uiTip>Web page summarising general downloads available for the software.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Other type of download for software - the default if a more specific type is not available.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="API specification"/> <xs:enumeration value="Biological data"/> <xs:enumeration value="Binaries"/> <xs:enumeration value="Command-line specification"/> <xs:enumeration value="Container file"/> <xs:enumeration value="Icon"/> <xs:enumeration value="Software package"/> <xs:enumeration value="Screenshot"/> <xs:enumeration value="Source code"/> <xs:enumeration value="Test data"/> <xs:enumeration value="Test script"/> <xs:enumeration value="Tool wrapper (CWL)"/> <xs:enumeration value="Tool wrapper (Galaxy)"/> <xs:enumeration value="Tool wrapper (Taverna)"/> <xs:enumeration value="Tool wrapper (Other)"/> <xs:enumeration value="VM image"/> <xs:enumeration value="Downloads page"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/download/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the download.</xs:documentation> <xs:appinfo> <uiTip>Comment about the download.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the download."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/download/version
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | versionType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="version" type="versionType" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this download.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/documentation
| diagram | 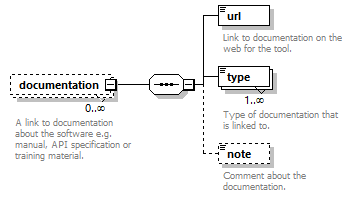 |
||||||
| namespace | biotoolsSchema | ||||||
| type | restriction of linkType | ||||||
| properties |
|
||||||
| children | url type note | ||||||
| annotation |
|
||||||
| source | <xs:element name="documentation" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A link to documentation about the software e.g. manual, API specification or training material.</xs:documentation> <xs:appinfo> <uiTip>A link to documentation about the software e.g. manual, API specification or training material.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:restriction base="linkType"> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>Link to documentation on the web for the tool.</xs:documentation> <xs:documentation> <uiTip>Documentation link.</uiTip> </xs:documentation> </xs:annotation> </xs:element> <xs:element name="type" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of documentation that is linked to.</xs:documentation> <xs:appinfo> <uiTip>Documentation type.</uiTip> <enum> <enumItem> <term>API documentation</term> <uiTip>Human-readable API documentation.</uiTip> </enumItem> <enumItem> <term>Citation instructions</term> <uiTip>Information on how to correctly cite use of the software; typically which publication(s) to cite, or something more general, e.g. a form of words to use.</uiTip> </enumItem> <enumItem> <term>Code of conduct</term> <uiTip>A set of guidelines or rules outlining the norms, expectations, responsibilities and proper practice for individuals working within the software project.</uiTip> </enumItem> <enumItem> <term>Command-line options</term> <uiTip>Information about the command-line interface to a tool.</uiTip> </enumItem> <enumItem> <term>Contributions policy</term> <uiTip>Information about policy for making contributions to the software project.</uiTip> </enumItem> <enumItem> <term>FAQ</term> <uiTip>Frequently Asked Questions (and answers) about the software.</uiTip> </enumItem> <enumItem> <term>General</term> <uiTip>General documentation.</uiTip> </enumItem> <enumItem> <term>Governance</term> <uiTip>Information about the software governance model.</uiTip> </enumItem> <enumItem> <term>Installation instructions</term> <uiTip>Instructions how to install the software.</uiTip> </enumItem> <enumItem> <term>Quick start guide</term> <uiTip>A short guide helping the end-user to use the software as soon as possible.</uiTip> </enumItem> <enumItem> <term>Release notes</term> <uiTip>Notes about a software release or changes to the software; a change log.</uiTip> </enumItem> <enumItem> <term>Terms of use</term> <uiTip>Rules that one must agree to abide by in order to use a service.</uiTip> </enumItem> <enumItem> <term>Training material</term> <uiTip>Online training material such as text on a Web page, a presentation, video etc. (but excluding tutorials).</uiTip> </enumItem> <enumItem> <term>User manual</term> <uiTip>Information on how to use the software, tailored to the end-user.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Some other type of documentation not listed in biotoolsSchema.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="API documentation"/> <xs:enumeration value="Citation instructions"/> <xs:enumeration value="Code of conduct"/> <xs:enumeration value="Command-line options"/> <xs:enumeration value="Contributions policy"/> <xs:enumeration value="FAQ"/> <xs:enumeration value="General"/> <xs:enumeration value="Governance"/> <xs:enumeration value="Installation instructions"/> <xs:enumeration value="Quick start guide"/> <xs:enumeration value="Release notes"/> <xs:enumeration value="Terms of use"/> <xs:enumeration value="Training material"/> <xs:enumeration value="User manual"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the documentation.</xs:documentation> <xs:appinfo> <uiTip>Comment about the documentation.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the documentation."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:restriction> </xs:complexContent> </xs:complexType> </xs:element> |
element tool/documentation/url
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | urlftpType | |||||||||
| properties |
|
|||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>Link to documentation on the web for the tool.</xs:documentation> <xs:documentation> <uiTip>Documentation link.</uiTip> </xs:documentation> </xs:annotation> </xs:element> |
element tool/documentation/type
| diagram |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of enumType | |||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="type" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of documentation that is linked to.</xs:documentation> <xs:appinfo> <uiTip>Documentation type.</uiTip> <enum> <enumItem> <term>API documentation</term> <uiTip>Human-readable API documentation.</uiTip> </enumItem> <enumItem> <term>Citation instructions</term> <uiTip>Information on how to correctly cite use of the software; typically which publication(s) to cite, or something more general, e.g. a form of words to use.</uiTip> </enumItem> <enumItem> <term>Code of conduct</term> <uiTip>A set of guidelines or rules outlining the norms, expectations, responsibilities and proper practice for individuals working within the software project.</uiTip> </enumItem> <enumItem> <term>Command-line options</term> <uiTip>Information about the command-line interface to a tool.</uiTip> </enumItem> <enumItem> <term>Contributions policy</term> <uiTip>Information about policy for making contributions to the software project.</uiTip> </enumItem> <enumItem> <term>FAQ</term> <uiTip>Frequently Asked Questions (and answers) about the software.</uiTip> </enumItem> <enumItem> <term>General</term> <uiTip>General documentation.</uiTip> </enumItem> <enumItem> <term>Governance</term> <uiTip>Information about the software governance model.</uiTip> </enumItem> <enumItem> <term>Installation instructions</term> <uiTip>Instructions how to install the software.</uiTip> </enumItem> <enumItem> <term>Quick start guide</term> <uiTip>A short guide helping the end-user to use the software as soon as possible.</uiTip> </enumItem> <enumItem> <term>Release notes</term> <uiTip>Notes about a software release or changes to the software; a change log.</uiTip> </enumItem> <enumItem> <term>Terms of use</term> <uiTip>Rules that one must agree to abide by in order to use a service.</uiTip> </enumItem> <enumItem> <term>Training material</term> <uiTip>Online training material such as text on a Web page, a presentation, video etc. (but excluding tutorials).</uiTip> </enumItem> <enumItem> <term>User manual</term> <uiTip>Information on how to use the software, tailored to the end-user.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>Some other type of documentation not listed in biotoolsSchema.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="API documentation"/> <xs:enumeration value="Citation instructions"/> <xs:enumeration value="Code of conduct"/> <xs:enumeration value="Command-line options"/> <xs:enumeration value="Contributions policy"/> <xs:enumeration value="FAQ"/> <xs:enumeration value="General"/> <xs:enumeration value="Governance"/> <xs:enumeration value="Installation instructions"/> <xs:enumeration value="Quick start guide"/> <xs:enumeration value="Release notes"/> <xs:enumeration value="Terms of use"/> <xs:enumeration value="Training material"/> <xs:enumeration value="User manual"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/documentation/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the documentation.</xs:documentation> <xs:appinfo> <uiTip>Comment about the documentation.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the documentation."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/relation
| diagram | 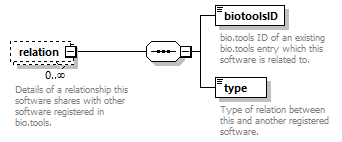 |
||||||
| namespace | biotoolsSchema | ||||||
| properties |
|
||||||
| children | biotoolsID type | ||||||
| annotation |
|
||||||
| source | <xs:element name="relation" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of a relationship this software shares with other software registered in bio.tools.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="biotoolsID" type="biotoolsIdType"> <xs:annotation> <xs:documentation>bio.tools ID of an existing bio.tools entry which this software is related to.</xs:documentation> <xs:documentation>Relations may only be defined between registered software. The ID is a URL in the bio.tools namespace and reflects (normally exactly) the tool name and version: see http://biotools.readthedocs.io/.</xs:documentation> <xs:appinfo> <uiTip>bio.tools ID (URL) of an existing bio.tools entry which this software is related to.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="type"> <xs:annotation> <xs:documentation>Type of relation between this and another registered software.</xs:documentation> <xs:documentation>Certain relations may only be defined between certain types of tool.</xs:documentation> <xs:appinfo> <uiTip>Type of relation between this and another registered software.</uiTip> <enum> <enumItem> <term>isNewVersionOf / hasNewVersion</term> <uiTip>The software is a new version of an existing software, typically providing new or improved functionality.</uiTip> </enumItem> <enumItem> <term>uses / usedBy</term> <uiTip>The software provides an interface to or in some other way uses the functions of other software under the hood,e.g. invoking a command-line tool or calling a Web API, Web service or SPARQL endpoint to perform its function.</uiTip> </enumItem> <enumItem> <term>includes / includedIn</term> <uiTip>A workbench, toolkit or workflow includes some other, independently available, software.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="isNewVersionOf"/> <xs:enumeration value="hasNewVersion"/> <xs:enumeration value="uses"/> <xs:enumeration value="usedBy"/> <xs:enumeration value="includes"/> <xs:enumeration value="includedIn"/> </xs:restriction> </xs:simpleType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element tool/relation/biotoolsID
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | biotoolsIdType | ||||||
| properties |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="biotoolsID" type="biotoolsIdType"> <xs:annotation> <xs:documentation>bio.tools ID of an existing bio.tools entry which this software is related to.</xs:documentation> <xs:documentation>Relations may only be defined between registered software. The ID is a URL in the bio.tools namespace and reflects (normally exactly) the tool name and version: see http://biotools.readthedocs.io/.</xs:documentation> <xs:appinfo> <uiTip>bio.tools ID (URL) of an existing bio.tools entry which this software is related to.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/relation/type
| diagram |  |
||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||
| source | <xs:element name="type"> <xs:annotation> <xs:documentation>Type of relation between this and another registered software.</xs:documentation> <xs:documentation>Certain relations may only be defined between certain types of tool.</xs:documentation> <xs:appinfo> <uiTip>Type of relation between this and another registered software.</uiTip> <enum> <enumItem> <term>isNewVersionOf / hasNewVersion</term> <uiTip>The software is a new version of an existing software, typically providing new or improved functionality.</uiTip> </enumItem> <enumItem> <term>uses / usedBy</term> <uiTip>The software provides an interface to or in some other way uses the functions of other software under the hood,e.g. invoking a command-line tool or calling a Web API, Web service or SPARQL endpoint to perform its function.</uiTip> </enumItem> <enumItem> <term>includes / includedIn</term> <uiTip>A workbench, toolkit or workflow includes some other, independently available, software.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="isNewVersionOf"/> <xs:enumeration value="hasNewVersion"/> <xs:enumeration value="uses"/> <xs:enumeration value="usedBy"/> <xs:enumeration value="includes"/> <xs:enumeration value="includedIn"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/publication
| diagram | 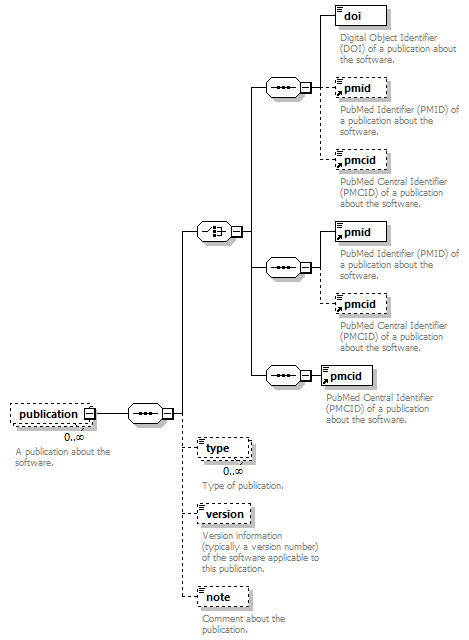 |
||||||
| namespace | biotoolsSchema | ||||||
| properties |
|
||||||
| children | doi pmid pmcid type version note | ||||||
| annotation |
|
||||||
| source | <xs:element name="publication" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>A publication about the software.</xs:documentation> <xs:appinfo> <uiTip>A publication about the software.</uiTip> > </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:choice> <xs:sequence> <xs:element name="doi" type="doiType"> <xs:annotation> <xs:appinfo> <uiTip>Digital Object Identifier.</uiTip> > </xs:appinfo> <xs:documentation>Digital Object Identifier (DOI) of a publication about the software.</xs:documentation> </xs:annotation> </xs:element> <xs:element ref="pmid" minOccurs="0"/> <xs:element ref="pmcid" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="pmid"/> <xs:element ref="pmcid" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="pmcid"/> </xs:sequence> </xs:choice> <xs:element name="type" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of publication. </xs:documentation> <xs:appinfo> <uiTip>Type of publication.</uiTip> <enum> <enumItem> <term>Primary</term> <uiTip>The principal publication about the tool itself; the article to cite when acknowledging use of the tool.</uiTip> </enumItem> <enumItem> <term>Method</term> <uiTip>A publication describing a scientific method or algorithm implemented by the tool.</uiTip> </enumItem> <enumItem> <term>Usage</term> <uiTip>A publication describing the application of the tool to scientific research, a particular task or dataset.</uiTip> </enumItem> <enumItem> <term>Benchmarking study</term> <uiTip>A publication which assessed the performance of the tool.</uiTip> </enumItem> <enumItem> <term>Review</term> <uiTip>A publication where the tool was reviewed.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>A publication of relevance to the tool but not fitting the other categories.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Primary"/> <xs:enumeration value="Benchmarking study"/> <xs:enumeration value="Method"/> <xs:enumeration value="Usage"/> <xs:enumeration value="Review"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="version" type="versionType" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this publication.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the publication.</xs:documentation> <xs:appinfo> <uiTip>Comment about the publication.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the publication."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element tool/publication/doi
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | doiType | ||||||
| properties |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="doi" type="doiType"> <xs:annotation> <xs:appinfo> <uiTip>Digital Object Identifier.</uiTip> > </xs:appinfo> <xs:documentation>Digital Object Identifier (DOI) of a publication about the software.</xs:documentation> </xs:annotation> </xs:element> |
element tool/publication/type
| diagram |  |
||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||
| source | <xs:element name="type" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of publication. </xs:documentation> <xs:appinfo> <uiTip>Type of publication.</uiTip> <enum> <enumItem> <term>Primary</term> <uiTip>The principal publication about the tool itself; the article to cite when acknowledging use of the tool.</uiTip> </enumItem> <enumItem> <term>Method</term> <uiTip>A publication describing a scientific method or algorithm implemented by the tool.</uiTip> </enumItem> <enumItem> <term>Usage</term> <uiTip>A publication describing the application of the tool to scientific research, a particular task or dataset.</uiTip> </enumItem> <enumItem> <term>Benchmarking study</term> <uiTip>A publication which assessed the performance of the tool.</uiTip> </enumItem> <enumItem> <term>Review</term> <uiTip>A publication where the tool was reviewed.</uiTip> </enumItem> <enumItem> <term>Other</term> <uiTip>A publication of relevance to the tool but not fitting the other categories.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Primary"/> <xs:enumeration value="Benchmarking study"/> <xs:enumeration value="Method"/> <xs:enumeration value="Usage"/> <xs:enumeration value="Review"/> <xs:enumeration value="Other"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/publication/version
| diagram |  |
|||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||
| type | versionType | |||||||||||||||
| properties |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:element name="version" type="versionType" minOccurs="0"> <xs:annotation> <xs:documentation>Version information (typically a version number) of the software applicable to this publication.</xs:documentation> <xs:appinfo> <uiTip>Software version number.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/publication/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the publication.</xs:documentation> <xs:appinfo> <uiTip>Comment about the publication.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the publication."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tool/credit
| diagram | 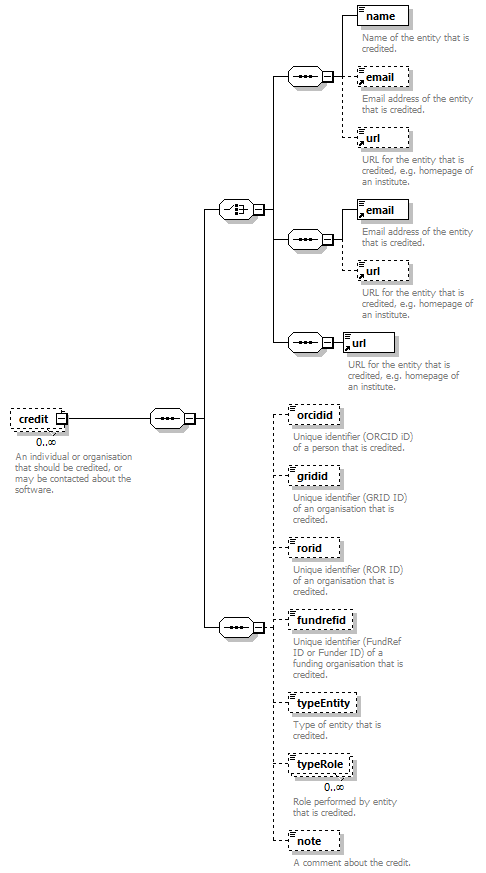 |
||||||
| namespace | biotoolsSchema | ||||||
| properties |
|
||||||
| children | name email url orcidid gridid rorid fundrefid typeEntity typeRole note | ||||||
| annotation |
|
||||||
| source | <xs:element name="credit" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>An individual or organisation that should be credited, or may be contacted about the software.</xs:documentation> <xs:appinfo> <uiTip>An individual or organisation that should be credited, or may be contacted about the software.</uiTip> </xs:appinfo> </xs:annotation> <xs:complexType> <xs:sequence> <xs:choice> <xs:sequence> <xs:element name="name"> <xs:annotation> <xs:appinfo> <uiTip>Name of credited entity.</uiTip> </xs:appinfo> <xs:documentation>Name of the entity that is credited.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Person or institute name"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:minLength value="1"/> <xs:maxLength value="100"/> <xs:whiteSpace value="collapse"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element ref="email" minOccurs="0"/> <xs:element ref="url" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="email"/> <xs:element ref="url" minOccurs="0"/> </xs:sequence> <xs:sequence> <xs:element ref="url"/> </xs:sequence> </xs:choice> <xs:sequence> <xs:element name="orcidid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>ORCID iD of person.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (ORCID iD) of a person that is credited.</xs:documentation> <xs:documentation>Open Researcher and Contributor IDs (ORCID IDs) provide a persistent reference to information on a researcher, see http://orcid.org/. </xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="https://orcid.org/0000-0002-1825-0097"/> <altova:example value="http://orcid.org/0000-0002-1825-009X"/> <altova:example value="https://orcid.org/0000-0002-1825-009X"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="http://orcid\.org/[0-9]{4}-[0-9]{4}-[0-9]{4}-[0-9]{3}[0-9X]"/> <xs:pattern value="https://orcid\.org/[0-9]{4}-[0-9]{4}-[0-9]{4}-[0-9]{3}[0-9X]"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="gridid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>GRID ID of organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (GRID ID) of an organisation that is credited. </xs:documentation> <xs:documentation>Global Research Identifier Database (GRID) IDs provide a persistent reference to information on research organisations, see https://www.grid.ac/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="grid.5170.3"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="grid.[0-9]{4,}.[a-f0-9]{1,2}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="rorid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>ROR ID of organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (ROR ID) of an organisation that is credited.</xs:documentation> <xs:documentation>Research Organization Registry (ROR) IDs provide a persistent reference to information on research organisations, see https://ror.org/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="03yrm5c26"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="0[0-9a-zA-Z]{6}[0-9]{2}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="fundrefid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>Crossref funder ID of a funding organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (FundRef ID or Funder ID) of a funding organisation that is credited.</xs:documentation> <xs:documentation>The Funder Registry (formerly FundRef) IDs provide a persistent reference to information on funding organisations registered in the Crossref registry, see https://www.crossref.org/services/funder-registry/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.13039/100009273"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="10\.13039/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="typeEntity" minOccurs="0"> <xs:annotation> <xs:documentation>Type of entity that is credited.</xs:documentation> <xs:appinfo> <uiTip>Type of entity that is credited.</uiTip> <enum> <enumItem> <term>Person</term> <uiTip>Credit of an individual.</uiTip> </enumItem> <enumItem> <term>Project</term> <uiTip>Credit of a community software project not formally associated with any single institute.</uiTip> </enumItem> <enumItem> <term>Division</term> <uiTip>Credit of or a formal part of an institutional organisation, e.g. a department, research group, team, etc</uiTip> </enumItem> <enumItem> <term>Institute</term> <uiTip>Credit of an organisation such as a university, hospital, research institute, service center, unit etc.</uiTip> </enumItem> <enumItem> <term>Consortium</term> <uiTip>Credit of an association of two or more institutes or other legal entities which have joined forces for some common purpose. Includes Research Infrastructures (RIs) such as ELIXIR, parts of an RI such as an ELIXIR node etc. </uiTip> </enumItem> <enumItem> <term>Funding agency</term> <uiTip>Credit of a legal entity providing funding for development of the software or provision of an online service.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Person"/> <xs:enumeration value="Project"/> <xs:enumeration value="Division"/> <xs:enumeration value="Institute"/> <xs:enumeration value="Consortium"/> <xs:enumeration value="Funding agency"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="typeRole" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:appinfo> <uiTip>Role performed by credited entity.</uiTip> <enum> <enumItem> <term>Developer</term> <uiTip>Author of the original software source code.</uiTip> </enumItem> <enumItem> <term>Maintainer</term> <uiTip>Maintainer of a mature software providing packaging, patching, distribution etc.</uiTip> </enumItem> <enumItem> <term>Provider</term> <uiTip>Institutional provider of an online service.</uiTip> </enumItem> <enumItem> <term>Documentor</term> <uiTip>Author of software documentation including making edits to a bio.tools entry.</uiTip> </enumItem> <enumItem> <term>Contributor</term> <uiTip>Some other role in software production or service delivery including design, deployment, system administration, evaluation, testing, documentation, training, user support etc.</uiTip> </enumItem> <enumItem> <term>Support</term> <uiTip>Provider of support in using the software.</uiTip> </enumItem> <enumItem> <term>Primary contact</term> <uiTip>The primary point of contact for the software.</uiTip> </enumItem> </enum> </xs:appinfo> <xs:documentation>Role performed by entity that is credited.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Primary contact"/> <xs:enumeration value="Contributor"/> <xs:enumeration value="Developer"/> <xs:enumeration value="Documentor"/> <xs:enumeration value="Maintainer"/> <xs:enumeration value="Provider"/> <xs:enumeration value="Support"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>A comment about the credit.</uiTip> </xs:appinfo> <xs:documentation>A comment about the credit.</xs:documentation> <xs:documentation>This can elaborate on the contribution of the credited entity.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the credit."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:sequence> </xs:complexType> </xs:element> |
element tool/credit/name
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | restriction of xs:token | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="name"> <xs:annotation> <xs:appinfo> <uiTip>Name of credited entity.</uiTip> </xs:appinfo> <xs:documentation>Name of the entity that is credited.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Person or institute name"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:minLength value="1"/> <xs:maxLength value="100"/> <xs:whiteSpace value="collapse"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/orcidid
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | restriction of xs:token | |||||||||
| properties |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="orcidid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>ORCID iD of person.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (ORCID iD) of a person that is credited.</xs:documentation> <xs:documentation>Open Researcher and Contributor IDs (ORCID IDs) provide a persistent reference to information on a researcher, see http://orcid.org/. </xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="https://orcid.org/0000-0002-1825-0097"/> <altova:example value="http://orcid.org/0000-0002-1825-009X"/> <altova:example value="https://orcid.org/0000-0002-1825-009X"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="http://orcid\.org/[0-9]{4}-[0-9]{4}-[0-9]{4}-[0-9]{3}[0-9X]"/> <xs:pattern value="https://orcid\.org/[0-9]{4}-[0-9]{4}-[0-9]{4}-[0-9]{3}[0-9X]"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/gridid
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:token | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="gridid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>GRID ID of organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (GRID ID) of an organisation that is credited. </xs:documentation> <xs:documentation>Global Research Identifier Database (GRID) IDs provide a persistent reference to information on research organisations, see https://www.grid.ac/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="grid.5170.3"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="grid.[0-9]{4,}.[a-f0-9]{1,2}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/rorid
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:token | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="rorid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>ROR ID of organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (ROR ID) of an organisation that is credited.</xs:documentation> <xs:documentation>Research Organization Registry (ROR) IDs provide a persistent reference to information on research organisations, see https://ror.org/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="03yrm5c26"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="0[0-9a-zA-Z]{6}[0-9]{2}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/fundrefid
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:token | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="fundrefid" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>Crossref funder ID of a funding organisation.</uiTip> </xs:appinfo> <xs:documentation>Unique identifier (FundRef ID or Funder ID) of a funding organisation that is credited.</xs:documentation> <xs:documentation>The Funder Registry (formerly FundRef) IDs provide a persistent reference to information on funding organisations registered in the Crossref registry, see https://www.crossref.org/services/funder-registry/.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.13039/100009273"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:token"> <xs:pattern value="10\.13039/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/typeEntity
| diagram |  |
||||||||||||||||||||||||
| namespace | biotoolsSchema | ||||||||||||||||||||||||
| type | restriction of enumType | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||
| source | <xs:element name="typeEntity" minOccurs="0"> <xs:annotation> <xs:documentation>Type of entity that is credited.</xs:documentation> <xs:appinfo> <uiTip>Type of entity that is credited.</uiTip> <enum> <enumItem> <term>Person</term> <uiTip>Credit of an individual.</uiTip> </enumItem> <enumItem> <term>Project</term> <uiTip>Credit of a community software project not formally associated with any single institute.</uiTip> </enumItem> <enumItem> <term>Division</term> <uiTip>Credit of or a formal part of an institutional organisation, e.g. a department, research group, team, etc</uiTip> </enumItem> <enumItem> <term>Institute</term> <uiTip>Credit of an organisation such as a university, hospital, research institute, service center, unit etc.</uiTip> </enumItem> <enumItem> <term>Consortium</term> <uiTip>Credit of an association of two or more institutes or other legal entities which have joined forces for some common purpose. Includes Research Infrastructures (RIs) such as ELIXIR, parts of an RI such as an ELIXIR node etc. </uiTip> </enumItem> <enumItem> <term>Funding agency</term> <uiTip>Credit of a legal entity providing funding for development of the software or provision of an online service.</uiTip> </enumItem> </enum> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Person"/> <xs:enumeration value="Project"/> <xs:enumeration value="Division"/> <xs:enumeration value="Institute"/> <xs:enumeration value="Consortium"/> <xs:enumeration value="Funding agency"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/typeRole
| diagram | 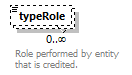 |
|||||||||||||||||||||||||||
| namespace | biotoolsSchema | |||||||||||||||||||||||||||
| type | restriction of enumType | |||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||
| source | <xs:element name="typeRole" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:appinfo> <uiTip>Role performed by credited entity.</uiTip> <enum> <enumItem> <term>Developer</term> <uiTip>Author of the original software source code.</uiTip> </enumItem> <enumItem> <term>Maintainer</term> <uiTip>Maintainer of a mature software providing packaging, patching, distribution etc.</uiTip> </enumItem> <enumItem> <term>Provider</term> <uiTip>Institutional provider of an online service.</uiTip> </enumItem> <enumItem> <term>Documentor</term> <uiTip>Author of software documentation including making edits to a bio.tools entry.</uiTip> </enumItem> <enumItem> <term>Contributor</term> <uiTip>Some other role in software production or service delivery including design, deployment, system administration, evaluation, testing, documentation, training, user support etc.</uiTip> </enumItem> <enumItem> <term>Support</term> <uiTip>Provider of support in using the software.</uiTip> </enumItem> <enumItem> <term>Primary contact</term> <uiTip>The primary point of contact for the software.</uiTip> </enumItem> </enum> </xs:appinfo> <xs:documentation>Role performed by entity that is credited.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="enumType"> <xs:enumeration value="Primary contact"/> <xs:enumeration value="Contributor"/> <xs:enumeration value="Developer"/> <xs:enumeration value="Documentor"/> <xs:enumeration value="Maintainer"/> <xs:enumeration value="Provider"/> <xs:enumeration value="Support"/> </xs:restriction> </xs:simpleType> </xs:element> |
element tool/credit/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:appinfo> <uiTip>A comment about the credit.</uiTip> </xs:appinfo> <xs:documentation>A comment about the credit.</xs:documentation> <xs:documentation>This can elaborate on the contribution of the credited entity.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="Comment about the credit."/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element tools
| diagram |  |
||
| namespace | biotoolsSchema | ||
| properties |
|
||
| children | tool | ||
| annotation |
|
||
| source | <xs:element name="tools"> <xs:annotation> <xs:documentation>Description of one or more bioinformatics tools - application software with well-defined data processing functions (inputs, outputs and operations). This includes simple tools with one or a few closely related functions, and complex, multimodal tools with many functions. Tools may be available available for immediate use as online services, or in a form which which you can download, install, configure and run yourself.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element ref="tool" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Description of a bioinformatics software tool - a software application with well-defined data processing functions (inputs, outputs and operations). A tool is a discrete, but possibly complex software entity.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element url
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | urlType | ||||||
| properties |
|
||||||
| used by |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="url" type="urlType"> <xs:annotation> <xs:documentation>URL for the entity that is credited, e.g. homepage of an institute.</xs:documentation> <xs:appinfo> <uiTip>URL for credited entity.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
complexType dataType
| diagram | 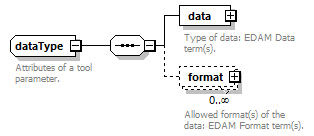 |
||
| namespace | biotoolsSchema | ||
| children | data format | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="dataType"> <xs:annotation> <xs:documentation>Attributes of a tool parameter.</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of data: EDAM Data term(s).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the data: EDAM Format term(s).</xs:documentation> <xs:appinfo> <uiTip>Allowed format(s) of the data: EDAM Format term(s).</uiTip> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
element dataType/data
| diagram | 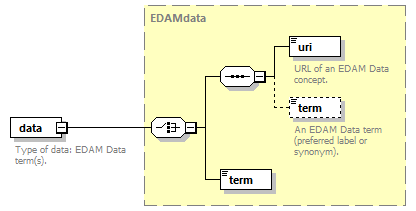 |
||
| namespace | biotoolsSchema | ||
| type | EDAMdata | ||
| properties |
|
||
| children | uri term term | ||
| annotation |
|
||
| source | <xs:element name="data" type="EDAMdata"> <xs:annotation> <xs:documentation>Type of data: EDAM Data term(s).</xs:documentation> </xs:annotation> </xs:element> |
element dataType/format
| diagram | 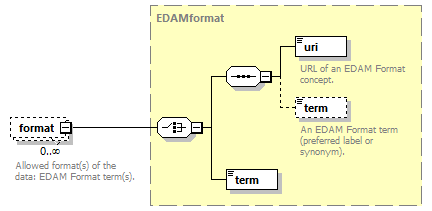 |
||||||
| namespace | biotoolsSchema | ||||||
| type | EDAMformat | ||||||
| properties |
|
||||||
| children | uri term term | ||||||
| annotation |
|
||||||
| source | <xs:element name="format" type="EDAMformat" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Allowed format(s) of the data: EDAM Format term(s).</xs:documentation> <xs:appinfo> <uiTip>Allowed format(s) of the data: EDAM Format term(s).</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
complexType EDAMdata
| diagram | 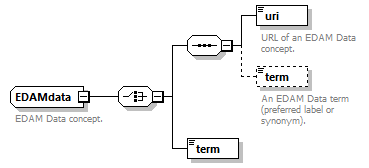 |
||
| namespace | biotoolsSchema | ||
| type | restriction of ontologyConcept | ||
| properties |
|
||
| children | uri term term | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="EDAMdata"> <xs:annotation> <xs:documentation>EDAM Data concept.</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Data concept.</xs:documentation> <xs:documentation>The URL must be in the EDAM Data namespace, i.e. http://edamontology.org/data_.</xs:documentation> <xs:appinfo> <uiTip>EDAM Data URL.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/data_2976"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/data_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Data term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Data term (preferred label or synonym).</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Protein sequences"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> |
element EDAMdata/uri
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:anyURI | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Data concept.</xs:documentation> <xs:documentation>The URL must be in the EDAM Data namespace, i.e. http://edamontology.org/data_.</xs:documentation> <xs:appinfo> <uiTip>EDAM Data URL.</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/data_2976"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/data_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element EDAMdata/term
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | xs:token | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Data term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Data term (preferred label or synonym).</uiTip> </xs:appinfo> <xs:appinfo> <altova:exampleValues> <altova:example value="Protein sequences"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> </xs:annotation> </xs:element> |
element EDAMdata/term
| diagram |  |
||
| namespace | biotoolsSchema | ||
| type | xs:token | ||
| properties |
|
||
| source | <xs:element name="term" type="xs:token"/> |
complexType EDAMformat
| diagram | 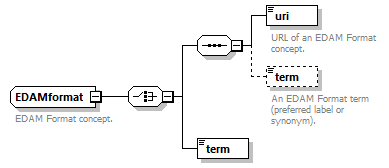 |
||
| namespace | biotoolsSchema | ||
| type | restriction of ontologyConcept | ||
| properties |
|
||
| children | uri term term | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="EDAMformat"> <xs:annotation> <xs:documentation>EDAM Format concept.</xs:documentation> </xs:annotation> <xs:complexContent> <xs:restriction base="ontologyConcept"> <xs:choice> <xs:sequence> <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Format concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Format URL.</uiTip> </xs:appinfo> <xs:documentation>The URL must be in the EDAM Format namespace, i.e. http://edamontology.org/format_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/format_1929"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/format_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Format term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Format term (preferred label or synonym)</uiTip> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="FASTA"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:restriction> </xs:complexContent> </xs:complexType> |
element EDAMformat/uri
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:anyURI | ||||||||
| properties |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="uri"> <xs:annotation> <xs:documentation>URL of an EDAM Format concept.</xs:documentation> <xs:appinfo> <uiTip>EDAM Format URL.</uiTip> </xs:appinfo> <xs:documentation>The URL must be in the EDAM Format namespace, i.e. http://edamontology.org/format_.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://edamontology.org/format_1929"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:anyURI"> <xs:pattern value="http://edamontology\.org/format_[0-9]{4}"/> </xs:restriction> </xs:simpleType> </xs:element> |
element EDAMformat/term
| diagram |  |
||||||||
| namespace | biotoolsSchema | ||||||||
| type | xs:token | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An EDAM Format term (preferred label or synonym).</xs:documentation> <xs:appinfo> <uiTip>EDAM Format term (preferred label or synonym)</uiTip> </xs:appinfo> <xs:documentation>The term must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="FASTA"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> </xs:element> |
element EDAMformat/term
| diagram |  |
||
| namespace | biotoolsSchema | ||
| type | xs:token | ||
| properties |
|
||
| source | <xs:element name="term" type="xs:token"/> |
complexType linkType
| diagram | 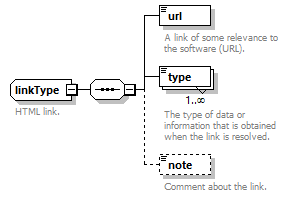 |
||
| namespace | biotoolsSchema | ||
| children | url type note | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="linkType"> <xs:annotation> <xs:documentation>HTML link.</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>A link of some relevance to the software (URL).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="type" type="enumType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The type of data or information that is obtained when the link is resolved.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the link.</xs:documentation> <xs:appinfo> <uiTip>Comment about the link.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
element linkType/url
| diagram |  |
|||||||||
| namespace | biotoolsSchema | |||||||||
| type | urlftpType | |||||||||
| properties |
|
|||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:element name="url" type="urlftpType"> <xs:annotation> <xs:documentation>A link of some relevance to the software (URL).</xs:documentation> </xs:annotation> </xs:element> |
element linkType/type
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | enumType | ||||||
| properties |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="type" type="enumType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The type of data or information that is obtained when the link is resolved.</xs:documentation> </xs:annotation> </xs:element> |
element linkType/note
| diagram |  |
||||||||||||
| namespace | biotoolsSchema | ||||||||||||
| type | textType | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="note" type="textType" minOccurs="0"> <xs:annotation> <xs:documentation>Comment about the link.</xs:documentation> <xs:appinfo> <uiTip>Comment about the link.</uiTip> </xs:appinfo> </xs:annotation> </xs:element> |
complexType ontologyConcept
| diagram | 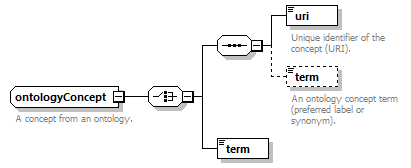 |
||||
| namespace | biotoolsSchema | ||||
| children | uri term term | ||||
| used by |
|
||||
| annotation |
|
||||
| source | <xs:complexType name="ontologyConcept"> <xs:annotation> <xs:documentation>A concept from an ontology.</xs:documentation> </xs:annotation> <xs:choice> <xs:sequence> <xs:element name="uri" type="xs:anyURI"> <xs:annotation> <xs:documentation>Unique identifier of the concept (URI).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An ontology concept term (preferred label or synonym).</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:element name="term" type="xs:token"/> </xs:choice> </xs:complexType> |
element ontologyConcept/uri
| diagram |  |
||
| namespace | biotoolsSchema | ||
| type | xs:anyURI | ||
| properties |
|
||
| annotation |
|
||
| source | <xs:element name="uri" type="xs:anyURI"> <xs:annotation> <xs:documentation>Unique identifier of the concept (URI).</xs:documentation> </xs:annotation> </xs:element> |
element ontologyConcept/term
| diagram |  |
||||||
| namespace | biotoolsSchema | ||||||
| type | xs:token | ||||||
| properties |
|
||||||
| annotation |
|
||||||
| source | <xs:element name="term" type="xs:token" minOccurs="0"> <xs:annotation> <xs:documentation>An ontology concept term (preferred label or synonym).</xs:documentation> </xs:annotation> </xs:element> |
element ontologyConcept/term
| diagram |  |
||
| namespace | biotoolsSchema | ||
| type | xs:token | ||
| properties |
|
||
| source | <xs:element name="term" type="xs:token"/> |
simpleType biotoolsIdType
| namespace | biotoolsSchema | ||||||||
| type | restriction of xs:anyURI | ||||||||
| properties |
|
||||||||
| used by |
|
||||||||
| facets |
|
||||||||
| annotation |
|
||||||||
| source | <xs:simpleType name="biotoolsIdType"> <xs:annotation> <xs:documentation>bio.tools tool ID.</xs:documentation> <xs:documentation>bio.tools tool IDs are a URL-safe and Linked-Data-safe derivative of (often identical to) the tool name. Allowed characters are uppercase and lowercase English letters (case insensitive!), decimal digits, hyphen, period, and underscore. Spaces can be preserved as underscore ("_").</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="needle"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>bio.tools tool IDs are set by bio.tools admin and will be disregarded if specified in a payload (e.g. PUSH, POST) to the bio.tools API.</xs:documentation> </xs:annotation> <xs:restriction base="xs:anyURI"> <xs:pattern value="[_\-.0-9a-zA-Z]*"/> </xs:restriction> </xs:simpleType> |
simpleType doiType
| namespace | biotoolsSchema | ||||||
| type | restriction of xs:token | ||||||
| properties |
|
||||||
| used by |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:simpleType name="doiType"> <xs:annotation> <xs:appinfo> <altova:exampleValues> <altova:example value="10.1093/nar/gkv1116"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>Digital Object Identifier.</xs:documentation> </xs:annotation> <xs:restriction base="xs:token"> <xs:pattern value="10\.[0-9]{4,9}/[\[\]<>A-Za-z0-9:;\)\(_/.-]+"/> </xs:restriction> </xs:simpleType> |
simpleType enumType
| namespace | biotoolsSchema | ||||||
| type | restriction of xs:string | ||||||
| properties |
|
||||||
| used by | |||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:simpleType name="enumType"> <xs:annotation> <xs:documentation>Enumeration of labels.</xs:documentation> </xs:annotation> <xs:restriction base="xs:string"> <xs:whiteSpace value="collapse"/> </xs:restriction> </xs:simpleType> |
simpleType nameType
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of xs:token | |||||||||||||||
| properties |
|
|||||||||||||||
| used by |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:simpleType name="nameType"> <xs:annotation> <xs:documentation>Tool or collection name.</xs:documentation> <xs:appinfo>The name has a 100 character limit and may only contain space, uppercase and lowercase letters, decimal digits, plus symbol, period, comma, dash, underscore, colon, semicolon and parentheses. Line feeds, carriage returns, tabs, leading and trailing spaces, and multiple spaces are not allowed / will be removed.</xs:appinfo> </xs:annotation> <xs:restriction base="xs:token"> <xs:minLength value="1"/> <xs:maxLength value="100"/> <xs:whiteSpace value="collapse"/> <xs:pattern value="[\p{Zs}A-Za-z0-9+\.,\-_:;()]*"/> </xs:restriction> </xs:simpleType> |
simpleType textType
| namespace | biotoolsSchema | ||||||||||||
| type | restriction of xs:token | ||||||||||||
| properties |
|
||||||||||||
| used by |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:simpleType name="textType"> <xs:annotation> <xs:documentation>Free text with a minimum length of 10 and maximum length of 1000. Line feeds, carriage returns, tabs, leading and trailing spaces, and multiple spaces are not allowed / will be removed.</xs:documentation> </xs:annotation> <xs:restriction base="xs:token"> <xs:whiteSpace value="collapse"/> <xs:maxLength value="1000"/> <xs:minLength value="10"/> </xs:restriction> </xs:simpleType> |
simpleType urlftpType
| namespace | biotoolsSchema | |||||||||
| type | restriction of xs:anyURI | |||||||||
| properties |
|
|||||||||
| used by |
|
|||||||||
| facets |
|
|||||||||
| annotation |
|
|||||||||
| source | <xs:simpleType name="urlftpType"> <xs:annotation> <xs:documentation>URL supporting FTP.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="http://someurl.org"/> <altova:example value="ftp://someurl.org"/> <altova:example value="https://someurl.org"/> <altova:example value="sftp://someurl.org"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:restriction base="xs:anyURI"> <xs:pattern value="http(s?)://[^\s/$.?#]*\.[^\s]*"/> <xs:pattern value="s?ftp://[^\s/$.?#]*\.[^\s]*"/> </xs:restriction> </xs:simpleType> |
simpleType urlType
| namespace | biotoolsSchema | ||||||
| type | restriction of xs:anyURI | ||||||
| properties |
|
||||||
| used by |
|
||||||
| facets |
|
||||||
| annotation |
|
||||||
| source | <xs:simpleType name="urlType"> <xs:annotation> <xs:documentation>An HTTP or HTTPS URL.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="https://someurl.org"/> <altova:example value="http://someurl.org"/> </altova:exampleValues> </xs:appinfo> </xs:annotation> <xs:restriction base="xs:anyURI"> <xs:pattern value="http(s?)://[^\s/$.?#]*\.[^\s]*"/> </xs:restriction> </xs:simpleType> |
simpleType versionType
| namespace | biotoolsSchema | |||||||||||||||
| type | restriction of xs:token | |||||||||||||||
| properties |
|
|||||||||||||||
| used by |
|
|||||||||||||||
| facets |
|
|||||||||||||||
| annotation |
|
|||||||||||||||
| source | <xs:simpleType name="versionType"> <xs:annotation> <xs:documentation>Tool version label or version range.</xs:documentation> <xs:appinfo> <altova:exampleValues> <altova:example value="1.1"/> <altova:example value="beta01"/> <altova:example value="2.0 - 2.7"/> <altova:example value="1.1, 1.2.1, 1.4, v5"/> <altova:example value="1.1 - 1.4, 2.0-alpha, 2.0-beta-01 - 2.0-beta-04, 2.0.0, 2.0~alpha-01"/> </altova:exampleValues> </xs:appinfo> <xs:documentation>The name has a 100 character limit and may only contain space, uppercase and lowercase English letters, decimal digits, plus symbol, period, comma, dash, colon, semicolon and parentheses.</xs:documentation> </xs:annotation> <xs:restriction base="xs:token"> <xs:minLength value="1"/> <xs:whiteSpace value="collapse"/> <xs:maxLength value="100"/> <xs:pattern value="[\p{Zs}A-Za-z0-9+\.,\-_:;()~]*"/> </xs:restriction> </xs:simpleType> |
XML Schema documentation generated by XMLSpy Schema Editor http://www.altova.com/xmlspy